User:Ashley Crotteau/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 41: | Line 41: | ||
===Cancer=== | ===Cancer=== | ||
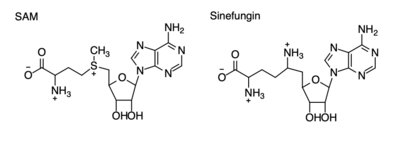
| - | Many studies have shown that lysine methyltransferases lead to the inhibition of cancers, therefore they are being studied as possible cancer therapy treatments. Lysine methylation contributes to the inactivation of a tumor suppressor gene. Due to this, KMTs are being studied as possible biomarkers for the detection of cancers. Inhibitors of KMT, such as Sinefungin (Figure 4), were used in a study to observe KMT’s effect on cancerous cells. It was found that when KMT is deregulated, tumor behavior increases due to a lack of methylation, indicating the importance of the histone methylation. <ref name=Xuejiao /> | + | Many studies have shown that lysine methyltransferases lead to the inhibition of cancers, therefore they are being studied as possible cancer therapy treatments. Lysine methylation contributes to the inactivation of a tumor suppressor gene. Due to this, KMTs are being studied as possible biomarkers for the detection of cancers. Inhibitors of KMT, such as Sinefungin (Figure 4), were used in a study to observe KMT’s effect on cancerous cells. It was found that when KMT is deregulated, tumor behavior increases due to a lack of methylation, indicating the importance of the histone methylation. <ref name=Xuejiao /> |
</StructureSection> | </StructureSection> | ||
Revision as of 15:12, 12 April 2019
H. sapiens Lysine Methyltransferase, SET 7/9
| |||||||||||
References
[3] [5] [1] [2] [4] [6] [7] [8]
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003 Jun;28(6):329-35. PMID:12826405
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Yeates TO. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002 Oct 4;111(1):5-7. PMID:12372294
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ 4.0 4.1 Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998 Jun 26;273(26):15933-9. PMID:9632640
- ↑ 5.0 5.1 doi: https://dx.doi.org/10.1016/C2014-0-02189-2
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Del Rizzo PA, Couture JF, Dirk LM, Strunk BS, Roiko MS, Brunzelle JS, Houtz RL, Trievel RC. SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation. J Biol Chem. 2010 Oct 8;285(41):31849-58. Epub 2010 Aug 1. PMID:20675860 doi:http://dx.doi.org/10.1074/jbc.M110.114587
- ↑ 7.0 7.1 7.2 7.3 7.4 Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010 Dec;21(12):2069-80. doi: 10.1681/ASN.2010060633. Epub 2010, Oct 7. PMID:20930066 doi:http://dx.doi.org/10.1681/ASN.2010060633
- ↑ 8.0 8.1 Tian X, Zhang S, Liu HM, Zhang YB, Blair CA, Mercola D, Sassone-Corsi P, Zi X. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: new targets for cancer therapy and prevention. Curr Cancer Drug Targets. 2013 Jun;13(5):558-79. doi:, 10.2174/1568009611313050007. PMID:23713993 doi:http://dx.doi.org/10.2174/1568009611313050007
Student Contributors
Ashley Crotteau
Parker Hiday
Lauren Allman