User:Karsten Theis/Insulin
From Proteopedia
(→Structure) |
|||
| Line 43: | Line 43: | ||
== Structure == | == Structure == | ||
<StructureSection load='' size='350' side='right' scene='82/821037/Ribbon/1' caption=''> | <StructureSection load='' size='350' side='right' scene='82/821037/Ribbon/1' caption=''> | ||
| - | The structure of insulin has been known for decades, but the structure of the receptor is not completely resolved yet. | ||
===Structure of mature insulin monomer=== | ===Structure of mature insulin monomer=== | ||
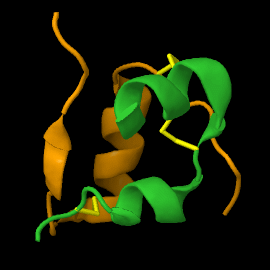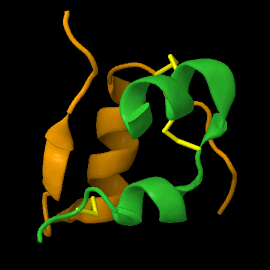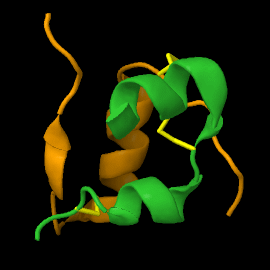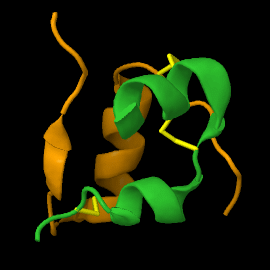
<scene name='82/821037/Ribbon/1'>Mature insulin</scene> contains two chains, A and B, held together by disulfide bonds and non-covalent interactions. The <scene name='82/821037/Spacefilling/2'>surface of insulin</scene> contains quite a few hydrophobic side chains, which form protein:protein contacts when insulin forms hexamers or binds to its receptor. Select coloring of the side chains below to explore the surface properties. | <scene name='82/821037/Ribbon/1'>Mature insulin</scene> contains two chains, A and B, held together by disulfide bonds and non-covalent interactions. The <scene name='82/821037/Spacefilling/2'>surface of insulin</scene> contains quite a few hydrophobic side chains, which form protein:protein contacts when insulin forms hexamers or binds to its receptor. Select coloring of the side chains below to explore the surface properties. | ||
| Line 66: | Line 65: | ||
</jmolRadioGroup> | </jmolRadioGroup> | ||
</jmol> | </jmol> | ||
| - | |||
| - | |||
| - | These two chains are joined by disulfide bonds, which are shown in yellow. | ||
===Targeting and processing=== | ===Targeting and processing=== | ||
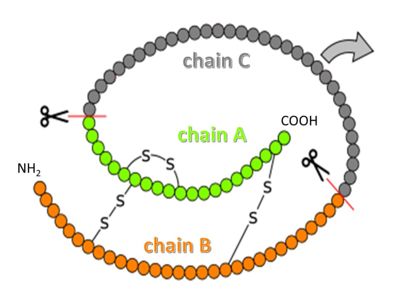
[[Image:Proinsulin.jpg|400 px]] | [[Image:Proinsulin.jpg|400 px]] | ||
| - | + | Insulin it synthesized as so-called preproinsulin and gets targeted into the ER and Golgi via a signal sequence, which then is removed to yield proinsulin. <scene name='82/821037/Proinsulin/1'>Proinsulin</scene> is processed by several proteases in the Golgi apparatus to form 3 separate chains, named B, C and A (see figure above)<ref>PMID:15289650</ref>. Chains B and A are linked through disulfide bonds, and are the components of mature insulin. | |
| - | <scene name='82/821037/Proinsulin/1'>Proinsulin</scene> | + | |
<jmol> | <jmol> | ||
| Line 95: | Line 90: | ||
<jmolLink> | <jmolLink> | ||
<script>anim mode loop; anim on</script> | <script>anim mode loop; anim on</script> | ||
| - | <text>disordered</text></jmolLink></jmol> in the proinsulin structure, is | + | <text>disordered</text></jmolLink></jmol> in the proinsulin structure, is no longer connected to insulin after processing and its fate and biological role after processing is unclear. |
| + | |||
===Storage=== | ===Storage=== | ||
Before insulin is secreted, it is able to pair-up with itself and form a dimer by forming hydrogen bonds between the ends of two B-chains, which then combine in threes to form a hexamer (a trimer of dimers to be exact). In crystal structures, insulin occurs in two states, <scene name='82/821037/Rvst/1'>T or R</scene>. In an animation (takes long to load), the <scene name='82/821037/Rvst/2'>dimer organisation is conserved in the R and the T state</scene>. | Before insulin is secreted, it is able to pair-up with itself and form a dimer by forming hydrogen bonds between the ends of two B-chains, which then combine in threes to form a hexamer (a trimer of dimers to be exact). In crystal structures, insulin occurs in two states, <scene name='82/821037/Rvst/1'>T or R</scene>. In an animation (takes long to load), the <scene name='82/821037/Rvst/2'>dimer organisation is conserved in the R and the T state</scene>. | ||
| - | (A) <scene name='82/821037/T6/1'>T6 hexamer</scene>, (B) T3Rf3 hexamer and (C) R6 hexamer. | + | (A) <scene name='82/821037/T6/1'>T6 hexamer (e.g. structure 4INS) </scene>, (B) T3Rf3 hexamer (e.g. 1TRZ) and (C) R6 hexamer (e.g. 1ZNJ). Small changes in the sequence of insulin change how fast hexamers fall apart into monomers. This is used to make insulin preparations that give off low levels of insulin for a longer time, or high levels of insulin for a short time when injected as microcrystals in depots under the skin. |
===Receptor interaction=== | ===Receptor interaction=== | ||
Revision as of 13:18, 13 July 2019
Insulin is a peptide hormone that helps to maintain blood sugar within a healthy range by regulating carbohydrate and lipid metabolism throughout the body[1][2]. It is secreted by specialized cells in the pancreas and acts by binding to insulin receptors on other cells. Insulin in its mature form contains two peptide chains connected by disulfide crosslinks, and occurs either as monomer or as hexamer. Administering insulin in carefully determined doses at the appropriate times is used in managing diabetes, a chronic condition where the body fails to maintain blood sugar levels by itself.
Contents |
Function
Insulin, together with glucagon, regulates blood sugar levels by changing fuel metabolism in all cells [3] on a timescale of minutes and hours. In simple terms, the presence of insulin in the blood signals the well-fed stage, while the presence of glucagon signals the fasting stage. [1]
- Biosynthesis
"The pancreas of a normal adult contains approximately 200 units of insulin, and the average daily secretion of insulin into the circulation in healthy individuals ranges from 30 to 50 units. https://www.britannica.com/science/insulin" 1 IU = 0.0347 mg
foo1[4]
- Secretion and transport
- Receptor interaction
foo3[6]
- Degradation
foo4[7]
Receptor recycling [8]
The body is able to sense the concentration of glucose in the blood and respond by secreting insulin, which is produced by beta cells in the pancreas. Insulin then binds to the insulin receptor, changing its conformation [9]. Depending on cell type and the presence of other signals (glucogon, epinephrine), the cell will modify its metabolism.
Disease and Treatment
In patients with diabetes, insulin signalling is compromised[10].
Type I: Insulin is not produced
Type II: Insulin is produced, but receptor does not trigger signal: [11]
Synthesis of human insulin in E. coli is important to producing insulin for the treatment of type 1 diabetes. It is believed that the hydrophobic sections on the B-chain cause insulin aggregation which initially caused problems in the manufacture and storage of insulin for pharmaceutical use.
In patients with diabetes, insulin signalling is compromised[13]. Synthesis of human insulin in E. coli is important to producing insulin for the treatment of type 1 diabetes. It is believed that the hydrophobic sections on the B-chain cause insulin aggregation which initially caused problems in the manufacture and storage of insulin for pharmaceutical use.
Structure
| |||||||||||
References
- ↑ Sonksen P, Sonksen J. Insulin: understanding its action in health and disease. Br J Anaesth. 2000 Jul;85(1):69-79. PMID:10927996
- ↑ Weiss MA, Lawrence MC. A thing of beauty: Structure and function of insulin's "aromatic triplet". Diabetes Obes Metab. 2018 Sep;20 Suppl 2:51-63. doi: 10.1111/dom.13402. PMID:30230175 doi:http://dx.doi.org/10.1111/dom.13402
- ↑ https://www.yourhormones.info/hormones/insulin/
- ↑ https://www.who.int/biologicals/expert_committee/BS_2143_Human_Recombinant_Insulin_final.pdf
- ↑ "https://www.diabetesselfmanagement.com/diabetes-resources/definitions/portal-vein/"
- ↑ https://pdb101.rcsb.org/motm/182
- ↑ Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998 Oct;19(5):608-24. doi: 10.1210/edrv.19.5.0349. PMID:9793760 doi:http://dx.doi.org/10.1210/edrv.19.5.0349
- ↑ Haeusler RA, McGraw TE, Accili D. Biochemical and cellular properties of insulin receptor signalling. Nat Rev Mol Cell Biol. 2018 Jan;19(1):31-44. doi: 10.1038/nrm.2017.89. Epub 2017 , Oct 4. PMID:28974775 doi:http://dx.doi.org/10.1038/nrm.2017.89
- ↑ Gutmann T, Kim KH, Grzybek M, Walz T, Coskun U. Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J Cell Biol. 2018 May 7;217(5):1643-1649. doi: 10.1083/jcb.201711047. Epub 2018, Feb 16. PMID:29453311 doi:http://dx.doi.org/10.1083/jcb.201711047
- ↑ https://www.endotext.org/section/diabetes/
- ↑ Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J Clin Invest. 2016 Jan;126(1):12-22. doi: 10.1172/JCI77812. Epub 2016 Jan 4. PMID:26727229 doi:http://dx.doi.org/10.1172/JCI77812
- ↑ doi: https://dx.doi.org/10.1530/endoabs.56.PL5
- ↑ https://www.endotext.org/section/diabetes/
- ↑ Davidson HW. (Pro)Insulin processing: a historical perspective. Cell Biochem Biophys. 2004;40(3 Suppl):143-58. PMID:15289650