This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
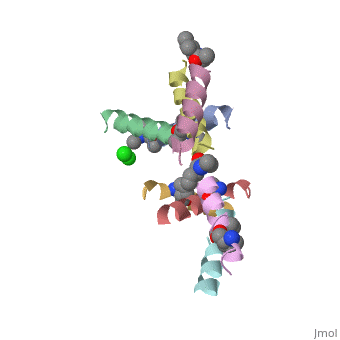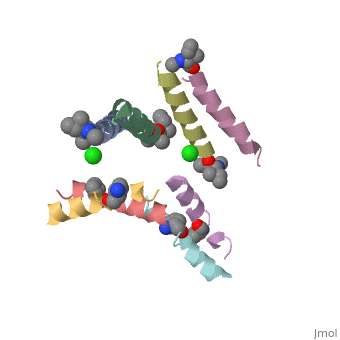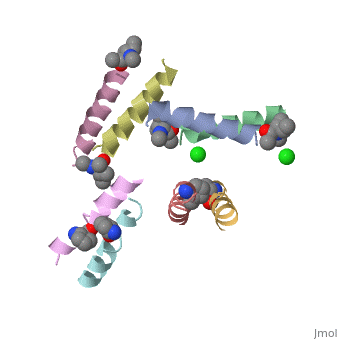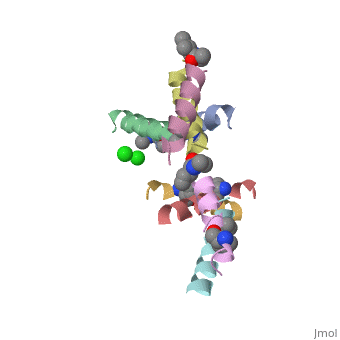
5cgn
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='5cgn' size='340' side='right'caption='[[5cgn]], [[Resolution|resolution]] 2.20Å' scene=''> | <StructureSection load='5cgn' size='340' side='right'caption='[[5cgn]], [[Resolution|resolution]] 2.20Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[5cgn]] is a 8 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5CGN OCA]. For a <b>guided tour on the structure components</b> use [ | + | <table><tr><td colspan='2'>[[5cgn]] is a 8 chain structure with sequence from [https://en.wikipedia.org/wiki/Synthetic_construct Synthetic construct]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5CGN OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=5CGN FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene> | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=DAL:D-ALANINE'>DAL</scene>, <scene name='pdbligand=DGL:D-GLUTAMIC+ACID'>DGL</scene>, <scene name='pdbligand=DHI:D-HISTIDINE'>DHI</scene>, <scene name='pdbligand=DIL:D-ISOLEUCINE'>DIL</scene>, <scene name='pdbligand=DLE:D-LEUCINE'>DLE</scene>, <scene name='pdbligand=DLY:D-LYSINE'>DLY</scene>, <scene name='pdbligand=DPN:D-PHENYLALANINE'>DPN</scene>, <scene name='pdbligand=DSE:N-METHYL-D-SERINE'>DSE</scene>, <scene name='pdbligand=DSG:D-ASPARAGINE'>DSG</scene>, <scene name='pdbligand=DVA:D-VALINE'>DVA</scene>, <scene name='pdbligand=MED:D-METHIONINE'>MED</scene>, <scene name='pdbligand=XCP:(1S,2S)-2-AMINOCYCLOPENTANECARBOXYLIC+ACID'>XCP</scene></td></tr> |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=5cgn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5cgn OCA], [https://pdbe.org/5cgn PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=5cgn RCSB], [https://www.ebi.ac.uk/pdbsum/5cgn PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=5cgn ProSAT]</span></td></tr> | |
| - | + | ||
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | |
</table> | </table> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/MAGA_XENLA MAGA_XENLA] Antimicrobial peptides that inhibit the growth of numerous species of bacteria and fungi and induce osmotic lysis of protozoa. Magainins are membrane lytic agents. | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
== Publication Abstract from PubMed == | == Publication Abstract from PubMed == | ||
| Line 26: | Line 26: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: Forest | + | [[Category: Synthetic construct]] |
| - | [[Category: Gellman | + | [[Category: Forest KT]] |
| - | [[Category: Hayouka | + | [[Category: Gellman SH]] |
| - | [[Category: Mortenson | + | [[Category: Hayouka Z]] |
| - | [[Category: Satyshur | + | [[Category: Mortenson DE]] |
| - | [[Category: Thomas | + | [[Category: Satyshur KA]] |
| - | [[Category: Weisblum | + | [[Category: Thomas NC]] |
| - | + | [[Category: Weisblum B]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 12:05, 14 June 2023
Structure of quasiracemic Ala-Magainin 2 with a beta amino acid substitution at position 8
| |||||||||||