We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox reserved 1560
From Proteopedia
(Difference between revisions)
| Line 32: | Line 32: | ||
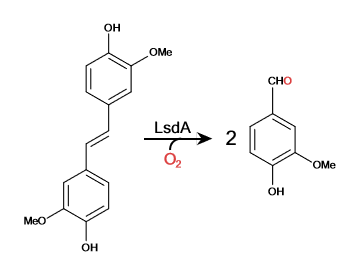
| - | Lignostilbene is a <scene name='83/830391/Chain_distingshin/2'>quationary</scene>structure with two identical | + | Lignostilbene is a <scene name='83/830391/Chain_distingshin/2'>quationary</scene> structure with two identical protein chains working together. The proteins are partially composed of <scene name='83/830391/Alpha_helix/1'>alpha helix's</scene>(Blue) and the majority by <scene name='83/830391/Beta_sheet/1'>Beta-sheets</scene> indicated in red. Together the <scene name='83/830391/Secoundary_structure_together/1'>tertiary</scene> structure work to form its relationship and interactions within itself and other bio-molecular compounds found in the environment. |
| + | |||
| + | A <scene name='82/823083/6ojttriad/1'>catalytic triad</scene> is commonly found in the active site of the protein and binds to the ligand. The triad for LsdA are identified as PHE 59, TYR 101, and LYS 134. The <scene name='83/830391/Ligand_nsl/1'>ligands</scene> are found within the two chains of the protein structure accompanied by an Iron ion. | ||
| - | A <scene name='82/823083/6ojttriad/1'>catalytic triad</scene> is commonly found in the active site of the and binds to the ligand, the proteins amino acids, PHE 59, TYR 101, and LYS 134 are identified as the triad. The <scene name='83/830391/Ligand_nsl/1'>ligands</scene> are found within the two chains of the protein in each Chain. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 01:18, 9 December 2019
| This Sandbox is Reserved from Aug 26 through Dec 12, 2019 for use in the course CHEM 351 Biochemistry taught by Bonnie_Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1556 through Sandbox Reserved 1575. |
To get started:
More help: Help:Editing |
Lignostilbene-α,β-dioxygenase A (LsdA) Catalyzation
| |||||||||||
References
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ https://pubchem.ncbi.nlm.nih.gov/compound/Vanillin
- ↑ Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol. 2009;60:165-82. doi: 10.1146/annurev.arplant.043008.092125. PMID:19014348 doi:http://dx.doi.org/10.1146/annurev.arplant.043008.092125