We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Johnson's Monday Lab Sandbox for Insulin Receptor
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
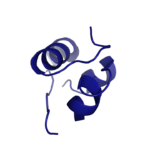
[[Image:Insulin.png|thumb|right|150px|Figure 1: Insulin molecule]] <scene name='83/839263/Insulin_molecule/1'>Insulin</scene> is a [http://en.wikipedia.org/wiki/Hormone hormone] that is synthesized and secreted from the [http://en.wikipedia.org/wiki/Pancreatic_islets islets of Langerhans] of the pancreas in response to high concentrations of glucose in the blood. Once it is secreted, insulin moves through the bloodstream and binds to unactivated insulin receptors residing in the plasma membrane. The receptor is fully activated after multiple insulin molecules are bound, and as previously mentioned, the regulation of various cellular processes is initiated. | [[Image:Insulin.png|thumb|right|150px|Figure 1: Insulin molecule]] <scene name='83/839263/Insulin_molecule/1'>Insulin</scene> is a [http://en.wikipedia.org/wiki/Hormone hormone] that is synthesized and secreted from the [http://en.wikipedia.org/wiki/Pancreatic_islets islets of Langerhans] of the pancreas in response to high concentrations of glucose in the blood. Once it is secreted, insulin moves through the bloodstream and binds to unactivated insulin receptors residing in the plasma membrane. The receptor is fully activated after multiple insulin molecules are bound, and as previously mentioned, the regulation of various cellular processes is initiated. | ||
==Structure== | ==Structure== | ||
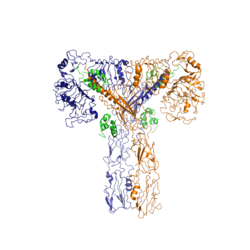
| - | The insulin receptor is a [http://en.wikipedia.org/wiki/Receptor_tyrosine_kinase receptor tyrosine kinase]. It is a heterotetramer that is constructed from two homodimers. Each homodimer maintains an extracellular domain, transmembrane helix, and an intracellular domain. The extracellular domain is divided into <scene name='83/839263/Alpha_and_beta_subunit/1'>alpha and beta</scene> [http://en.wikipedia.org/wiki/Protein_subunit | + | The insulin receptor is a [http://en.wikipedia.org/wiki/Receptor_tyrosine_kinase receptor tyrosine kinase]. It is a heterotetramer that is constructed from two homodimers. Each homodimer maintains an extracellular domain, transmembrane helix, and an intracellular domain. The extracellular domain is divided into <scene name='83/839263/Alpha_and_beta_subunit/1'>alpha and beta</scene> [http://en.wikipedia.org/wiki/Protein_subunit subunits]. The alpha subunit is characterized by two leucine-rich regions and one cysteine-rich region. The beta subunit contains three fibronectin type III domains. The alpha and beta subunits of the extracellular domains fold over one another and form a <scene name='83/839263/V_shape/1'>"V" shape</scene> when the insulin receptor is inactivated. Upon activation, the extracellular domain undergoes a conformational change and forms a <scene name='83/839263/T-shape/2'>"T" shape</scene>. |
[[Image:Insulin Receptor T.png|thumb|right|250px|Figure 2: Insulin receptor in the active "T" shape conformation with four insulins bound]] | [[Image:Insulin Receptor T.png|thumb|right|250px|Figure 2: Insulin receptor in the active "T" shape conformation with four insulins bound]] | ||
| - | An additional component to the [http://en.wikipedia.org/wiki/Ectodomain ectodomain] is the ''alpha'' chain C-terminal helix <ref name= "Uchikawa" />. The ''alpha''-CT is a single alpha-helix and it plays an important role in insulin binding and stabilization of the "T" shape activated conformation. The ''alpha''-CT interacts with a leucine-rich region of the alpha subunit and a fibronectin type III region of the beta subunit to form | + | An additional component to the [http://en.wikipedia.org/wiki/Ectodomain ectodomain] is the ''alpha'' chain C-terminal helix, which is also referred to as the "''alpha''-CT" <ref name= "Uchikawa" />. The ''alpha''-CT is a single alpha-helix and it plays an important role in insulin binding and stabilization of the "T" shape activated conformation. The ''alpha''-CT interacts with a leucine-rich region of the alpha subunit and a fibronectin type III region of the beta subunit to form the insulin binding sites known as site 1 and site 1' <ref name="Uchikawa" />. |
The structure of the extracellular domain is stabilized through [http://en.wikipedia.org/wiki/Covalent_bond covalent bonds]. The alpha subunits are linked through two disulfide bonds. Cys468 and <scene name='83/839263/Cys_holding_alphas_together/1'>Cys524</scene> of one alpha subunit are bound to Cys435 and <scene name='83/839263/Cys_holding_alphas_together/1'>Cys524</scene> of the other alpha subunit, respectively <ref name="Schäffer" />. <scene name='83/839263/Cys_683_holding_alphas_togethe/1'>Cys683</scene> of both alpha subunits hold the two together with a disulfide bond <ref name="Sparrow" />. The alpha subunit is held to the beta subunit by a disulfide bond between the <scene name='83/839263/Alpha_beta_link_by_disulfide/1'>Cys647 of the alpha subunit and Cys872 of the beta subunit</scene><ref name="Sparrow" /> | The structure of the extracellular domain is stabilized through [http://en.wikipedia.org/wiki/Covalent_bond covalent bonds]. The alpha subunits are linked through two disulfide bonds. Cys468 and <scene name='83/839263/Cys_holding_alphas_together/1'>Cys524</scene> of one alpha subunit are bound to Cys435 and <scene name='83/839263/Cys_holding_alphas_together/1'>Cys524</scene> of the other alpha subunit, respectively <ref name="Schäffer" />. <scene name='83/839263/Cys_683_holding_alphas_togethe/1'>Cys683</scene> of both alpha subunits hold the two together with a disulfide bond <ref name="Sparrow" />. The alpha subunit is held to the beta subunit by a disulfide bond between the <scene name='83/839263/Alpha_beta_link_by_disulfide/1'>Cys647 of the alpha subunit and Cys872 of the beta subunit</scene><ref name="Sparrow" /> | ||
| - | The insulin receptor extends intracellularly from the beta subunits of the ectodomain by way of a [http://en.wikipedia.org/wiki/Transmembrane_protein transmembrane] helix. Intracellularly, the insulin receptor contains two tyrosine kinase domains. | + | The insulin receptor extends intracellularly from the beta subunits of the ectodomain by way of a [http://en.wikipedia.org/wiki/Transmembrane_protein transmembrane] helix. Intracellularly, the insulin receptor contains two tyrosine kinase domains. |
===Insulin Binding=== | ===Insulin Binding=== | ||
| - | The insulin receptor unit has four separate sites for the insulin | + | The insulin receptor unit has four separate sites for the insulin binding. There are two pairs of two identical binding sites referred to as sites 1 and 1' and sites 2 and 2'. The insulin molecules bind to these sites mostly through [http://en.wikipedia.org/wiki/Hydrophobic_effect hydrophobic interactions], with some of the most crucial residues at sites 1 and 1' being between <scene name='83/839263/Residues_of_site_1_binding/4'>Cys A7, Cys B7, and His B5 of insulin and Pro495, Phe497, and Arg498</scene> of the insulin receptor FnIII-1 domain <ref name="Uchikawa" />. At sites 2 and 2', the major residues contributing to these hydrophobic interactions are the <scene name='83/839263/Site_2_residues_hydrophobic/1'>Leu 486, Leu 552, and Pro537 of the insulin receptor and Leu A13, Try A14, Leu A16, Leu B6, Ala B14, Leu B17 and Val B18 of the insulin molecule</scene><ref name="Uchikawa" />. While the majority of the binding interactions appear similar, sites 1 and 1' have a higher binding affinity than sites 2 and 2' due to site 1 having a larger surface area (706 square angstroms) exposed for insulin to bind to compared to site 2 (394 square angstroms)<ref name="Uchikawa" />. The binding interactions of the insulin molecules in sites 1 and 1' are facilitated by an <scene name='83/839263/Insulin_bound_to_site_1/2'>alpha helix</scene> of the insulin receptor. The insulin molecules in sites 2 and 2' primarily interact with the residues that comprise some of the <scene name='83/839263/Insulin_in_site_2_with_beta_sh/5'>beta-sheets</scene> of the insulin receptor. |
| - | + | At least three insulin molecules have to bind to the insulin receptor to induce the active "T-state" conformation <ref name="Uchikawa" />. The difference between the fully bound state with four insulins and the three-insulin-bound state is minimal compared to the difference between two and three insulins bound <ref name="Uchikawa" />. However, binding only two insulin molecules is insufficient to move the receptor to the active "T-state". | |
===Conformational Changes=== | ===Conformational Changes=== | ||
Revision as of 00:45, 31 March 2020
Insulin Receptor
| |||||||||||