Sulfotransferase
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Structural highlights == | == Structural highlights == | ||
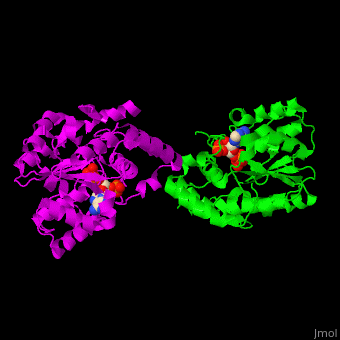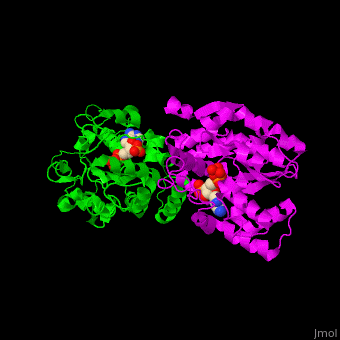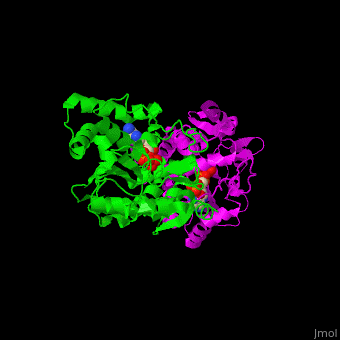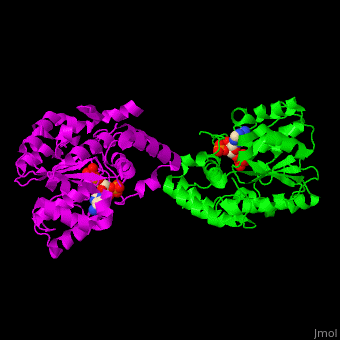
ST <scene name='49/491888/Cv/9'>catalytic residues His and Lys interact with the sulfate donor molecule PAP</scene><ref>PMID:11884392</ref>. <scene name='49/491888/Cv/10'>Whole active site</scene>. Water molecules are shown as red spheres. | ST <scene name='49/491888/Cv/9'>catalytic residues His and Lys interact with the sulfate donor molecule PAP</scene><ref>PMID:11884392</ref>. <scene name='49/491888/Cv/10'>Whole active site</scene>. Water molecules are shown as red spheres. | ||
| + | |||
| + | ==3D structures of sulfotransferase== | ||
| + | [[Sulfotransferase 3D structures]] | ||
</StructureSection> | </StructureSection> | ||
==3D structures of sulfotransferase== | ==3D structures of sulfotransferase== | ||
| Line 48: | Line 51: | ||
**[[1zrh]] - hST + PAP<BR /> | **[[1zrh]] - hST + PAP<BR /> | ||
| - | **[[3bd9]] - hST (mutant) + PAP<BR /> | + | **[[3bd9]], [[7scd]], [[7sce]] - hST (mutant) + PAP<BR /> |
**[[1t8u]] - hST catalytic domain + tetrasaccharide + PAP<BR /> | **[[1t8u]] - hST catalytic domain + tetrasaccharide + PAP<BR /> | ||
**[[3uan]] - mST + pentasaccharide + PAP<BR /> | **[[3uan]] - mST + pentasaccharide + PAP<BR /> | ||
| Line 127: | Line 130: | ||
**[[1t8t]] - hST catalytic domain + PAP<BR /> | **[[1t8t]] - hST catalytic domain + PAP<BR /> | ||
| + | **[[6xkg]], [[6xl8]] - hST catalytic domain + oligosaccharide<BR /> | ||
*''Sulfotransferase 4A1'' | *''Sulfotransferase 4A1'' | ||
| Line 140: | Line 144: | ||
**[[2zyu]] - mST + p-nitrophenyl sulfate + PAP<BR /> | **[[2zyu]] - mST + p-nitrophenyl sulfate + PAP<BR /> | ||
**[[2zvq]] - mST + α-naphthol + PAP<BR /> | **[[2zvq]] - mST + α-naphthol + PAP<BR /> | ||
| + | |||
| + | *''Sulfotransferase 2A8'' | ||
| + | |||
| + | **[[7d1x]] - mST + ADP<BR /> | ||
| + | **[[7eov]] - mST + cholic acid + PAP<BR /> | ||
*''Sulfotransferase 7A1'' | *''Sulfotransferase 7A1'' | ||
Revision as of 08:03, 13 April 2022
| |||||||||||
3D structures of sulfotransferase
Updated on 13-April-2022
References
- ↑ Malojcic G, Glockshuber R. The PAPS-independent aryl sulfotransferase and the alternative disulfide bond formation system in pathogenic bacteria. Antioxid Redox Signal. 2010 Oct;13(8):1247-59. doi: 10.1089/ars.2010.3119. PMID:20136513 doi:http://dx.doi.org/10.1089/ars.2010.3119
- ↑ Yalcin EB, More V, Neira KL, Lu ZJ, Cherrington NJ, Slitt AL, King RS. Downregulation of sulfotransferase expression and activity in diseased human livers. Drug Metab Dispos. 2013 Sep;41(9):1642-50. doi: 10.1124/dmd.113.050930. Epub 2013, Jun 17. PMID:23775849 doi:http://dx.doi.org/10.1124/dmd.113.050930
- ↑ Pedersen LC, Petrotchenko E, Shevtsov S, Negishi M. Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction. J Biol Chem. 2002 May 17;277(20):17928-32. Epub 2002 Mar 7. PMID:11884392 doi:http://dx.doi.org/10.1074/jbc.M111651200