We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1639
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
== Important amino acids == | == Important amino acids == | ||
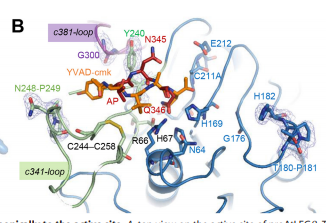
| - | To dig deeper and better understand the protein, you can see where the <scene name='86/861621/Protein_view_2/3'>ligand is bound to the protein</scene>. However, these ligands aren't entirely that important to the active site. The ligands shown are there to help stabilize the structure. The actual important enzyme of the active site is actually the activation peptide(AP). This AP [[Image:AP.PNG]] is made up of amino acid residues of ASN345 (asparagine), GLN346 (glutamine), ARG347 (arginine), which form<scene name='86/861621/Catalytic_triad/1'> the catalytic triad</scene> and where the cleavage happens. This cleavage [[Image:Cleavage.png]] basically splits the sequence in half of prime and nonprime on either side of the split. Then, the split sequences then take on a cyclic formation. | + | To dig deeper and better understand the protein, you can see where the <scene name='86/861621/Protein_view_2/3'>ligand is bound to the protein</scene>. However, these ligands aren't entirely that important to the active site. The ligands shown are there to help stabilize the structure. The actual important enzyme of the active site is actually the activation peptide(AP). This AP [[Image:AP.PNG]] is made up of amino acid residues of ASN345 (asparagine), GLN346 (glutamine), ARG347 (arginine), which form<scene name='86/861621/Catalytic_triad/1'> the catalytic triad</scene> and where the cleavage happens. This cleavage [[Image:Cleavage.png]] basically splits the sequence in half of prime and nonprime on either side of the split. Then, the split sequences then take on a cyclic formation. This cyclic formation is crucial for the protein to remain in homeostasis. |
Our protein is also very <scene name='86/861621/Hydrophobic_parts/1'>hydrophobic at certain ends.</scene> [[Image:Hydrophobic.png]] | Our protein is also very <scene name='86/861621/Hydrophobic_parts/1'>hydrophobic at certain ends.</scene> [[Image:Hydrophobic.png]] | ||
== Structural highlights == | == Structural highlights == | ||
Revision as of 16:56, 8 December 2020
| This Sandbox is Reserved from 09/18/2020 through 03/20/2021 for use in CHEM 351 Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, IA. This reservation includes Sandbox Reserved 1628 through Sandbox Reserved 1642. |
To get started:
More help: Help:Editing |
Arabidopsis thaliana legumain isoform beta in zymogen state (6YSA)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
[1] PMID:32719006