We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1656
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
== Structure == | == Structure == | ||
| - | <ref>PMID: | + | It is the catalytic domain which defines the family of the DUB. Indeed, DUBs belonging to the family of cysteine proteases have a catalytic site composed of two or three amino acids (dyads or triads). When the catalytic site is active, it may contain cysteine, histidine, aspartate or asparagine residues. In the case of metalloproteases, the active site is composed of a zinc ion and amino acids such as histidine, aspartate and serine. <ref>https://authors.library.caltech.edu/261/1/AMBpb04.pdf</ref> |
| + | |||
| + | Residues present in the catalytic site of DUBs are often in a non-functional orientation when the substrate is absent. Thus, when the substrate binds to the catalytic site of the enzyme, the site undergoes rearrangement and takes on a functional conformation. <ref>PMID:16537382</ref> | ||
== Biological role == | == Biological role == | ||
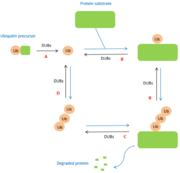
| - | The role of DUBs is in the ubiquitin pathway. The modifications made by DUBs are post-translational modifications. Thus, DUBs have different functions related to ubiquitin: A : maturation of ubiquitin, B : cleavage between protein and mono-ubiquitin and regulation of the poly-ubiquitin chain, C : cleavage between protein and poly-ubiquitin chain, D : recycling of ubiquitin. <ref>PMID:15571815</ref> | + | The role of DUBs is in the ubiquitin pathway. The modifications made by DUBs are post-translational modifications. Thus, DUBs have different functions related to ubiquitin: |
| + | |||
| + | A : maturation of ubiquitin, | ||
| + | |||
| + | B : cleavage between protein and mono-ubiquitin and regulation of the poly-ubiquitin chain, | ||
| + | |||
| + | C : cleavage between protein and poly-ubiquitin chain, | ||
| + | |||
| + | D : recycling of ubiquitin. <ref>PMID:15571815</ref> | ||
| Line 36: | Line 46: | ||
== Disease == | == Disease == | ||
| - | The involvement of deubiquitinases in diseases is still poorly understood. However, it is known that they play a role in various physiological processes, particularly in the case of cancers. <ref>PMID: | + | The involvement of deubiquitinases in diseases is still poorly understood. However, it is known that they play a role in various physiological processes, particularly in the case of cancers. <ref>PMID:19007433</ref> |
In fact, DUBs have a role in the mechanism involved in histone modification and so have influence on tumor development and progression. For instance, in gastric cancer, DUBs are regulated upwards and DUBs are related to tumor size. <ref>PMID:31897112</ref> | In fact, DUBs have a role in the mechanism involved in histone modification and so have influence on tumor development and progression. For instance, in gastric cancer, DUBs are regulated upwards and DUBs are related to tumor size. <ref>PMID:31897112</ref> | ||
Revision as of 21:22, 10 January 2021
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
Deubiquitinase
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997 Dec;11(14):1245-56. PMID:9409543
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Urbe S, Liu H, Hayes SD, Heride C, Rigden DJ, Clague MJ. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol Biol Cell. 2012 Mar;23(6):1095-103. doi: 10.1091/mbc.E11-08-0668. Epub 2012, Feb 1. PMID:22298430 doi:http://dx.doi.org/10.1091/mbc.E11-08-0668
- ↑ https://authors.library.caltech.edu/261/1/AMBpb04.pdf
- ↑ Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006 Mar 21;103(12):4675-80. Epub 2006 Mar 13. PMID:16537382
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem. 2008 Oct 21;9 Suppl 1:S3. doi: 10.1186/1471-2091-9-S1-S3. PMID:19007433 doi:http://dx.doi.org/10.1186/1471-2091-9-S1-S3
- ↑ Sun J, Shi X, Mamun MAA, Gao Y. The role of deubiquitinating enzymes in gastric cancer. Oncol Lett. 2020 Jan;19(1):30-44. doi: 10.3892/ol.2019.11062. Epub 2019 Nov 7. PMID:31897112 doi:http://dx.doi.org/10.3892/ol.2019.11062