We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1656
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
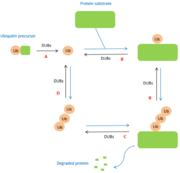
Deubiquitinases belong to the protease family. This family is divided into five classes, according to the nature of the amino acid composition of their active site carrying out the catalysis: serine protease, cysteine proteases, acid proteases, metalloproteases, threonine proteases. DUBs belong to only two of these families: metalloproteases and cysteine proteases. | Deubiquitinases belong to the protease family. This family is divided into five classes, according to the nature of the amino acid composition of their active site carrying out the catalysis: serine protease, cysteine proteases, acid proteases, metalloproteases, threonine proteases. DUBs belong to only two of these families: metalloproteases and cysteine proteases. | ||
| - | Among the cysteine proteins, four subfamilies can be described according to their catalytic domains: ubiquitin-specific proteases (USP), les Ubiquitin C-terminal hydrolases (UCH), Otubain proteases (OTU) and Machado-joseph disease proteases (MJD). The deubiquitinases belonging to the family of metalloproteases all have a JAMM catalytic domain (JAB1/MPN/Mov34 metalloenzyme). <ref>PMID:15571815</ref> | + | Among the cysteine proteins, four subfamilies can be described according to their catalytic domains: ubiquitin-specific proteases (USP), les Ubiquitin C-terminal hydrolases (UCH), Otubain proteases (OTU) and Machado-joseph disease proteases (MJD). The deubiquitinases belonging to the family of metalloproteases all have a JAMM catalytic domain (JAB1/MPN/Mov34 metalloenzyme). |
| + | Within these two families, DUBs are classified into subfamilies according to the differences in their amino acid sequences surrounding the catalytically active amino acid residues. <ref>PMID:15571815</ref> | ||
==== Localization ==== | ==== Localization ==== | ||
Revision as of 17:42, 11 January 2021
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
Deubiquitinase
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997 Dec;11(14):1245-56. PMID:9409543
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Urbe S, Liu H, Hayes SD, Heride C, Rigden DJ, Clague MJ. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol Biol Cell. 2012 Mar;23(6):1095-103. doi: 10.1091/mbc.E11-08-0668. Epub 2012, Feb 1. PMID:22298430 doi:http://dx.doi.org/10.1091/mbc.E11-08-0668
- ↑ https://authors.library.caltech.edu/261/1/AMBpb04.pdf
- ↑ Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006 Mar 21;103(12):4675-80. Epub 2006 Mar 13. PMID:16537382
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem. 2008 Oct 21;9 Suppl 1:S3. doi: 10.1186/1471-2091-9-S1-S3. PMID:19007433 doi:http://dx.doi.org/10.1186/1471-2091-9-S1-S3
- ↑ Sun J, Shi X, Mamun MAA, Gao Y. The role of deubiquitinating enzymes in gastric cancer. Oncol Lett. 2020 Jan;19(1):30-44. doi: 10.3892/ol.2019.11062. Epub 2019 Nov 7. PMID:31897112 doi:http://dx.doi.org/10.3892/ol.2019.11062
[1] Ubiquitine https://fr.wikipedia.org/wiki/Ubiquitine
[2] Ubiquitination https://fr.wikipedia.org/wiki/Ubiquitination