We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1656
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
==== Function ==== | ==== Function ==== | ||
| - | Deubiquitinases or Deubiquitinating enzymes (DUBs) are key enzymes belonging to the vast group of proteases, allowing the degradation of [https://fr.wikipedia.org/wiki/Ubiquitine ubiquitin] of proteins. These enzymes are thus implicated in the regulation of protein degradation. Indeed, when a protein is going to be degraded, an enzymatic cascade will add a poly-ubiquitin fragment to the protein. This mechanism is called [https://fr.wikipedia.org/wiki/Ubiquitination ubiquitination]. Following this step, mono or poly-ubiquitin is removed from the protein which has been degraded, by deubiquitinase. <ref>PMID:9409543</ref> | + | Deubiquitinases or Deubiquitinating enzymes (DUBs) are key enzymes belonging to the vast group of '''proteases''', allowing the degradation of [https://fr.wikipedia.org/wiki/Ubiquitine ubiquitin] of proteins. These enzymes are thus implicated in the regulation of '''protein degradation'''. Indeed, when a protein is going to be degraded, an enzymatic cascade will add a poly-ubiquitin fragment to the protein. This mechanism is called [https://fr.wikipedia.org/wiki/Ubiquitination ubiquitination]. Following this step, mono or poly-ubiquitin is removed from the protein which has been degraded, by deubiquitinase. <ref>PMID:9409543</ref> |
==== Families ==== | ==== Families ==== | ||
| - | Deubiquitinases belong to the protease family. This family is divided into five classes, according to the nature of the amino acid composition of their active site carrying out the catalysis: serine protease, [https://fr.wikipedia.org/wiki/Prot%C3%A9ase_%C3%A0_cyst%C3%A9ine cysteine proteases], acid proteases, metalloproteases, threonine proteases. DUBs belong to only two of these families: metalloproteases and cysteine proteases. | + | Deubiquitinases belong to the protease family. This family is divided into '''five classes''', according to the nature of the amino acid composition of their active site carrying out the catalysis: serine protease, [https://fr.wikipedia.org/wiki/Prot%C3%A9ase_%C3%A0_cyst%C3%A9ine cysteine proteases], acid proteases, metalloproteases, threonine proteases. DUBs belong to only two of these families: '''metalloproteases''' and '''cysteine proteases'''. |
Among the cysteine proteins, four subfamilies can be described according to their catalytic domains: ubiquitin-specific proteases (USP), les Ubiquitin C-terminal hydrolases (UCH), Otubain proteases (OTU) and Machado-joseph disease proteases (MJD). The deubiquitinases belonging to the family of metalloproteases all have a JAMM catalytic domain (JAB1/MPN/Mov34 metalloenzyme). | Among the cysteine proteins, four subfamilies can be described according to their catalytic domains: ubiquitin-specific proteases (USP), les Ubiquitin C-terminal hydrolases (UCH), Otubain proteases (OTU) and Machado-joseph disease proteases (MJD). The deubiquitinases belonging to the family of metalloproteases all have a JAMM catalytic domain (JAB1/MPN/Mov34 metalloenzyme). | ||
Within these two families, DUBs are classified into subfamilies according to the differences in their amino acid sequences surrounding the catalytically active amino acid residues. <ref>PMID:15571815</ref> | Within these two families, DUBs are classified into subfamilies according to the differences in their amino acid sequences surrounding the catalytically active amino acid residues. <ref>PMID:15571815</ref> | ||
| Line 24: | Line 24: | ||
==== Catalytic domain ==== | ==== Catalytic domain ==== | ||
| - | It is the catalytic domain which defines the family of the DUB. Indeed, DUBs belonging to the family of cysteine proteases have a catalytic site composed of two or three amino acids (dyads or triads). When the catalytic site is active, it may contain cysteine, histidine, aspartate or asparagine residues. In the case of metalloproteases, the active site is composed of a zinc ion and amino acids such as histidine, aspartate and serine. <ref>https://authors.library.caltech.edu/261/1/AMBpb04.pdf</ref> | + | It is the '''catalytic domain''' which defines the family of the DUB. Indeed, DUBs belonging to the family of cysteine proteases have a catalytic site composed of two or three amino acids (dyads or triads). When the catalytic site is active, it may contain cysteine, histidine, aspartate or asparagine residues. In the case of metalloproteases, the active site is composed of a zinc ion and amino acids such as histidine, aspartate and serine. <ref>https://authors.library.caltech.edu/261/1/AMBpb04.pdf</ref> |
| - | Residues present in the catalytic site of DUBs are often in a non-functional orientation when the substrate is absent. Thus, when the substrate binds to the catalytic site of the enzyme, the site undergoes rearrangement and takes on a functional conformation. <ref>PMID:16537382</ref> | + | Residues present in the catalytic site of DUBs are often in a '''non-functional orientation''' when the substrate is absent. Thus, when the substrate binds to the catalytic site of the enzyme, the site undergoes rearrangement and takes on a functional conformation. <ref>PMID:16537382</ref> |
== Biological role == | == Biological role == | ||
| Line 32: | Line 32: | ||
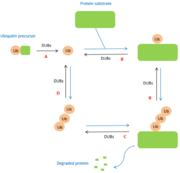
The role of DUBs is in the ubiquitin pathway. The modifications made by DUBs are post-translational modifications. Thus, DUBs have different functions related to ubiquitin: | The role of DUBs is in the ubiquitin pathway. The modifications made by DUBs are post-translational modifications. Thus, DUBs have different functions related to ubiquitin: | ||
| - | A : maturation of ubiquitin. When ubiquitin molecules are synthesized, they are not in free form. Thus, DUBs are essential for the generation of free monomers from precursors. The degradation of precursors is carried out by several DUBs belonging to the USPs class. | + | A : '''maturation of ubiquitin'''. When ubiquitin molecules are synthesized, they are not in free form. Thus, DUBs are essential for the generation of free monomers from precursors. The degradation of precursors is carried out by several DUBs belonging to the USPs class. |
B : cleavage between protein and mono-ubiquitin and regulation of the poly-ubiquitin chain. DUBs also have a regulatory activity because they allow the elimination of ubiquitin chains mistakenly conjugated to substrates. | B : cleavage between protein and mono-ubiquitin and regulation of the poly-ubiquitin chain. DUBs also have a regulatory activity because they allow the elimination of ubiquitin chains mistakenly conjugated to substrates. | ||
| Line 38: | Line 38: | ||
C : cleavage between protein and poly-ubiquitin chain. Once the protein has been degraded by the proteasome or autophagolysosome, DUBs allow the polyubiquitin chain to be separated from the protein. | C : cleavage between protein and poly-ubiquitin chain. Once the protein has been degraded by the proteasome or autophagolysosome, DUBs allow the polyubiquitin chain to be separated from the protein. | ||
| - | D : recycling of ubiquitin. Certain deubiquitinases allow the separation of polyubiquitin chains in order to release ubiquitin monomers. <ref>PMID:15571815</ref> | + | D : '''recycling of ubiquitin'''. Certain deubiquitinases allow the separation of polyubiquitin chains in order to release ubiquitin monomers. <ref>PMID:15571815</ref> |
Revision as of 21:47, 13 January 2021
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
Deubiquitinase
| |||||||||||
References
- ↑ Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007 Jan 12;315(5809):201-5. doi: 10.1126/science.1127085. PMID:17218518 doi:http://dx.doi.org/10.1126/science.1127085
- ↑ Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003 Sep 19;278(38):35857-60. doi: 10.1074/jbc.R300018200. Epub 2003, Jul 14. PMID:12860974 doi:http://dx.doi.org/10.1074/jbc.R300018200
- ↑ Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997 Dec;11(14):1245-56. PMID:9409543
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Urbe S, Liu H, Hayes SD, Heride C, Rigden DJ, Clague MJ. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol Biol Cell. 2012 Mar;23(6):1095-103. doi: 10.1091/mbc.E11-08-0668. Epub 2012, Feb 1. PMID:22298430 doi:http://dx.doi.org/10.1091/mbc.E11-08-0668
- ↑ https://authors.library.caltech.edu/261/1/AMBpb04.pdf
- ↑ Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006 Mar 21;103(12):4675-80. Epub 2006 Mar 13. PMID:16537382
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem. 2008 Oct 21;9 Suppl 1:S3. doi: 10.1186/1471-2091-9-S1-S3. PMID:19007433 doi:http://dx.doi.org/10.1186/1471-2091-9-S1-S3
- ↑ Sun J, Shi X, Mamun MAA, Gao Y. The role of deubiquitinating enzymes in gastric cancer. Oncol Lett. 2020 Jan;19(1):30-44. doi: 10.3892/ol.2019.11062. Epub 2019 Nov 7. PMID:31897112 doi:http://dx.doi.org/10.3892/ol.2019.11062
- ↑ Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer. 2019 Jan 1;26(1):R1-R14. doi: 10.1530/ERC-18-0264. PMID:30400005 doi:http://dx.doi.org/10.1530/ERC-18-0264
[1] Ubiquitine https://fr.wikipedia.org/wiki/Ubiquitine
[2] Ubiquitination https://fr.wikipedia.org/wiki/Ubiquitination
[3] Cysteine protease https://fr.wikipedia.org/wiki/Prot%C3%A9ase_%C3%A0_cyst%C3%A9ine
[4] Microtubule https://fr.wikipedia.org/wiki/Microtubule