We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Induced fit
From Proteopedia
(Difference between revisions)
(→Interactive examples) |
(→Interactive examples) |
||
| Line 16: | Line 16: | ||
<jmolRadioGroup> | <jmolRadioGroup> | ||
<item> | <item> | ||
| - | <script>anim off;model | + | <script>anim off;model 1</script> |
<text>Open</text> | <text>Open</text> | ||
</item> | </item> | ||
<item> | <item> | ||
| - | <script>anim off;model | + | <script>anim off;model 11</script> |
<text>Closed</text> | <text>Closed</text> | ||
</item> | </item> | ||
Current revision
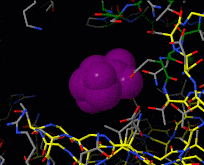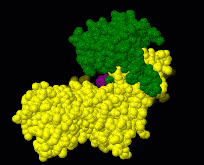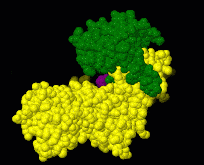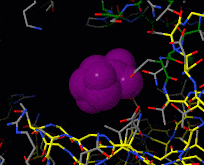
Induced fit describes a conformational change in a protein when it binds a ligand, in contrast to a lock-and-key model of ligand binding. A classic example of induced fit is binding of glucose to hexokinase, depicted in a morph between 3o8m and 3o80 in the picture at right.
Contents |
History of the concept
Induced fit was suggested by Koshland in 1958 [1], providing an alternative to the lock-and-key binding model that Emil Fischer proposed in 1899 [2].
Interactive examples
| |||||||||||
See Also
- A morph of induced fit when Tamiflu binds to Neuraminidase.