Journal:Acta Cryst D:S205979832000501X
From Proteopedia
(Difference between revisions)

| Line 12: | Line 12: | ||
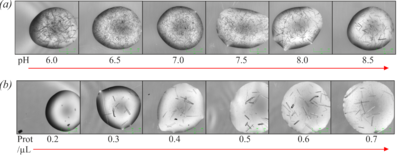
[[Image:Fig5ab.png|left|400px|thumb|'''Figure 5''' (a) Optimisation of crystallisation pH using bis-tris-propane buffer. (b) Optimisation of protein concentration.]] | [[Image:Fig5ab.png|left|400px|thumb|'''Figure 5''' (a) Optimisation of crystallisation pH using bis-tris-propane buffer. (b) Optimisation of protein concentration.]] | ||
| - | <scene name='84/842878/Cv/2'>Crystal structure of GBA dimer obtained at 1.56 Å resolution</scene> (PDB [[6tjk]]). ''N''-glycans are shown in spacefill representation. <scene name='84/842878/Cv/3'>GBA monomer comprising of three domains</scene>. Domain I (residues 1-27 and 383-414) in red, domain II (residues 30-75 and 431-497) in blue and domain III (residues 76-381 and 416-430) in gold. <scene name='84/842878/Cv/5'>Active site</scene> structure of bound bis-tris-propane which forms hydrogen bonds with Trp179, Asn234, Glu235, Glu340, Trp381 and an ethylene glycol (EDO) cryoprotectant molecules. <scene name='84/842878/Cv/6'>Overlay of recombinant GBA (gold) obtained at pH 7.0 and Cerezyme (cyan) obtained at pH 4.6</scene> (PDB [[6tjj]]). | + | <scene name='84/842878/Cv/2'>Crystal structure of GBA dimer in complex with bis-Tris propane (BTP) obtained at 1.56 Å resolution</scene> (PDB [[6tjk]]). ''N''-glycans are shown in spacefill representation. <scene name='84/842878/Cv/3'>GBA monomer comprising of three domains</scene>. Domain I (residues 1-27 and 383-414) in red, domain II (residues 30-75 and 431-497) in blue and domain III (residues 76-381 and 416-430) in gold. <scene name='84/842878/Cv/5'>Active site</scene> structure of bound bis-tris-propane which forms hydrogen bonds with Trp179, Asn234, Glu235, Glu340, Trp381 and an ethylene glycol (EDO) cryoprotectant molecules. <scene name='84/842878/Cv/6'>Overlay of recombinant GBA (gold) obtained at pH 7.0 and Cerezyme (cyan) obtained at pH 4.6</scene> (PDB [[6tjj]]). |
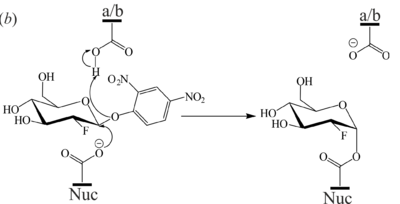
<scene name='84/842878/Cv/8'>Active site structure of the 2-deoxy-2-fluoro-β-D-glucopyranoside-GBA covalent intermediate</scene> (PDB [[6tjq]]). The 2F-Glc moiety is covalently bound to the catalytic nucleophile (Glu340) which occupies two conformations. a/b = catalytic acid-base, Nuc = catalytic nucleophile, EDO = ethylene glycol. | <scene name='84/842878/Cv/8'>Active site structure of the 2-deoxy-2-fluoro-β-D-glucopyranoside-GBA covalent intermediate</scene> (PDB [[6tjq]]). The 2F-Glc moiety is covalently bound to the catalytic nucleophile (Glu340) which occupies two conformations. a/b = catalytic acid-base, Nuc = catalytic nucleophile, EDO = ethylene glycol. | ||
[[Image:Fig7b.png|left|400px|thumb|'''Figure 7''' (b) Mechanism of 2F-DNPGlc hydrolysis by GBA to generate the covalent glycosyl-enzyme intermediate.]] | [[Image:Fig7b.png|left|400px|thumb|'''Figure 7''' (b) Mechanism of 2F-DNPGlc hydrolysis by GBA to generate the covalent glycosyl-enzyme intermediate.]] | ||
| - | <scene name='84/842878/Cv/9'>Crystal structure of GBA monomer obtained at 0.98 Å resolution</scene> (PDB [[6tn1]]). Domain I (residues 1-27 and 383-414) in orange, domain II (residues 30-75 and 431-497) in violet and domain III (residues 76–381 and 416-430) in blue. ''N''-glycans are shown in ball-and-stick representation. <scene name='84/842878/Cv/11'>Overlay of GBA unliganded structure with | + | <scene name='84/842878/Cv/9'>Crystal structure of GBA monomer obtained at 0.98 Å resolution</scene> (PDB [[6tn1]]). Domain I (residues 1-27 and 383-414) in orange, domain II (residues 30-75 and 431-497) in violet and domain III (residues 76–381 and 416-430) in blue. ''N''-glycans are shown in ball-and-stick representation. <scene name='84/842878/Cv/11'>Overlay of GBA unliganded structure with BTP complexed structure</scene> (PDB [[6tjk]]). Red indicates areas of high RMSD between the protein backbones. Loop 1 contains residues 27-31, loop 2 comprises residues 314-319 and loop 3 contains residues 344-350. <scene name='84/842878/Cv/13'>Active site of GBA unliganded crystal structure (blue) overlaid with active site residues of the BTP complex structure (gold) (PDB 6TJK) and Cerezyme (green) (PDB 6TJJ)</scene>. A magnesium ion (peach) coordinated by four waters (grey), Glu340 (nuc) and Glu235 (a/b), occupies the active site. |
<b>References</b><br> | <b>References</b><br> | ||
Revision as of 18:48, 27 June 2021
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.