This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1692
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
Amino Acids <scene name='89/892735/Highlighting_amino_acids/2'>85, 150, 166, 170, 202, 220, 275, and 331</scene> provide important interactions for binding. The <scene name='89/892735/His85_and_his105/1'>His105 side chain forms a hydrogen bond with His85</scene> side chain, providing stabilizing assistance. <ref name="Kondo">PMID:34303708</ref>. | Amino Acids <scene name='89/892735/Highlighting_amino_acids/2'>85, 150, 166, 170, 202, 220, 275, and 331</scene> provide important interactions for binding. The <scene name='89/892735/His85_and_his105/1'>His105 side chain forms a hydrogen bond with His85</scene> side chain, providing stabilizing assistance. <ref name="Kondo">PMID:34303708</ref>. | ||
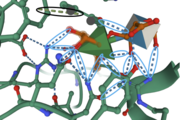
Amino Acid Residues <scene name='89/892735/Tyr150_and_his85/1'>Tyr150 and His85</scene> form a hydrogen bond with the O-1 atom of Rha, suggesting these residues play a catalytic role for the elimination reaction. [[Image:IntermolforcesBIOCHEM.png|thumb]] | Amino Acid Residues <scene name='89/892735/Tyr150_and_his85/1'>Tyr150 and His85</scene> form a hydrogen bond with the O-1 atom of Rha, suggesting these residues play a catalytic role for the elimination reaction. [[Image:IntermolforcesBIOCHEM.png|thumb]] | ||
| + | The image to the right shows important amino acids in the active site. Hydrogen bonding and pi-stacking interactions are indicated by the blue and black circles, respectively. | ||
== Structural highlights == | == Structural highlights == | ||
Secondary Structure: In this enzyme, there are around <scene name='89/892735/Beta_sheets/3'>30 anti-parallel beta sheets</scene> and three small <scene name='89/892735/Helices/1'>alpha helices</scene>. The anti-parallel beta sheets provide further stabilization, through strong hydrogen bonding in the backbone, of the enzyme compared to parallel beta sheets. The hydrophobic alpha helices provide structure for the formation of the active site. The structure of FoRham1 consists of a seven-bladed, Beta sheet-propellor domain that contributes to the shape and overall interactions with the active site, which is located on the anterior side of the enzyme. Here, <scene name='89/892735/His48/1'>His48</scene> acts as a catalytic residue that aids in the binding of GA into the active site. | Secondary Structure: In this enzyme, there are around <scene name='89/892735/Beta_sheets/3'>30 anti-parallel beta sheets</scene> and three small <scene name='89/892735/Helices/1'>alpha helices</scene>. The anti-parallel beta sheets provide further stabilization, through strong hydrogen bonding in the backbone, of the enzyme compared to parallel beta sheets. The hydrophobic alpha helices provide structure for the formation of the active site. The structure of FoRham1 consists of a seven-bladed, Beta sheet-propellor domain that contributes to the shape and overall interactions with the active site, which is located on the anterior side of the enzyme. Here, <scene name='89/892735/His48/1'>His48</scene> acts as a catalytic residue that aids in the binding of GA into the active site. | ||
Revision as of 02:38, 8 December 2021
| This Sandbox is Reserved from 10/01/2021 through 01/01//2022 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1690 through Sandbox Reserved 1699. |
To get started:
More help: Help:Editing |
Structure and Function of FoRham1
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Kondo T, Kichijo M, Maruta A, Nakaya M, Takenaka S, Arakawa T, Fushinobu S, Sakamoto T. Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family. J Biol Chem. 2021 Jul 22:101001. doi: 10.1016/j.jbc.2021.101001. PMID:34303708 doi:http://dx.doi.org/10.1016/j.jbc.2021.101001