We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1694
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
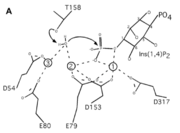
Inositol (1,4)-bisphosphate is a <scene name='89/892737/Substrate/2'>substrate</scene> that binds in the active site of INPP1D54A. The phosphate (1-PO4) in this substrate serves as a ligand for both the calcium ions in the mutated version of INPP1. <ref name="dollins">PMID:33172890</ref> The substrate is held in place by certain <scene name='89/892737/Aa_rotating/1'>amino acids</scene> (Glu269, Gly290, Lys294, Ala291, Ser157, Thr158) that play an extensive role in interactions such as hydrogen and ionic bonds between 1-PO4 and 2-PO4 of the substrate to the active site of the enzyme. Also, there are a few different ones <scene name='89/892737/Aa_for_ring/1'>Thr312, Lys270, Ala291, Asp156</scene> that interact with the inositol ring and help reinforce the binding of the Ins(1,4)P2 by possible hydrophobic, hydrogen, and ionic bonds. | Inositol (1,4)-bisphosphate is a <scene name='89/892737/Substrate/2'>substrate</scene> that binds in the active site of INPP1D54A. The phosphate (1-PO4) in this substrate serves as a ligand for both the calcium ions in the mutated version of INPP1. <ref name="dollins">PMID:33172890</ref> The substrate is held in place by certain <scene name='89/892737/Aa_rotating/1'>amino acids</scene> (Glu269, Gly290, Lys294, Ala291, Ser157, Thr158) that play an extensive role in interactions such as hydrogen and ionic bonds between 1-PO4 and 2-PO4 of the substrate to the active site of the enzyme. Also, there are a few different ones <scene name='89/892737/Aa_for_ring/1'>Thr312, Lys270, Ala291, Asp156</scene> that interact with the inositol ring and help reinforce the binding of the Ins(1,4)P2 by possible hydrophobic, hydrogen, and ionic bonds. | ||
| - | |||
| - | |||
Revision as of 14:57, 9 December 2021
| This Sandbox is Reserved from 10/01/2021 through 01/01//2022 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1690 through Sandbox Reserved 1699. |
To get started:
More help: Help:Editing |
Inositol polyphosphate 1-Phosphatase (INPP1) D54A
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 3.2 Dollins DE, Xiong JP, Endo-Streeter S, Anderson DE, Bansal VS, Ponder JW, Ren Y, York JD. A Structural Basis for Lithium and Substrate Binding of an Inositide Phosphatase. J Biol Chem. 2020 Nov 10. pii: RA120.014057. doi: 10.1074/jbc.RA120.014057. PMID:33172890 doi:http://dx.doi.org/10.1074/jbc.RA120.014057