User:Mathias Bortoletto Dunker/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 69: | Line 69: | ||
<scene name='89/898348/Nrf2dom3/1'>Neh3</scene>, <scene name='89/898348/Nrf2dom4/1'>Neh4</scene> and <scene name='89/898348/Nrf2dom5/1'>Neh5</scene> domain (red, blue and light blue): may play a role in NRF2 protein stability and may act as a transactivation domain, interacting with component of the transcriptional apparatus (Neh3) or to a protein called cAMP Response Element Binding Protein (Neh4 and Neh5), which possesses intrinsic histone acetyltransferase activity. | <scene name='89/898348/Nrf2dom3/1'>Neh3</scene>, <scene name='89/898348/Nrf2dom4/1'>Neh4</scene> and <scene name='89/898348/Nrf2dom5/1'>Neh5</scene> domain (red, blue and light blue): may play a role in NRF2 protein stability and may act as a transactivation domain, interacting with component of the transcriptional apparatus (Neh3) or to a protein called cAMP Response Element Binding Protein (Neh4 and Neh5), which possesses intrinsic histone acetyltransferase activity. | ||
| - | Neh6 domain (yellow): may contain a degron that is involved in a redox-insensitive process of degradation of NRF2. This occurs even in stressed cells, which normally extend the half-life of NRF2 protein relative to unstressed conditions by suppressing other degradation pathways. | + | <scene name='89/898348/Nrf2dom6/2'>Neh6 domain</scene>(yellow): may contain a degron that is involved in a redox-insensitive process of degradation of NRF2. This occurs even in stressed cells, which normally extend the half-life of NRF2 protein relative to unstressed conditions by suppressing other degradation pathways. |
| - | Nh7 domain (green): | + | <scene name='89/898348/Nrf2dom7/1'>Nh7 domain </scene>(green): |
The gene responsible for Nrf2 coding in humans is called HAGRID 283, and its sequence of bases is available in the [https://genomics.senescence.info/genes/entry.php?hgnc=NFE2L2 Human Ageing Genomic Resources] database, and it is as follows: | The gene responsible for Nrf2 coding in humans is called HAGRID 283, and its sequence of bases is available in the [https://genomics.senescence.info/genes/entry.php?hgnc=NFE2L2 Human Ageing Genomic Resources] database, and it is as follows: | ||
[[Image:Nrf2gene.jpg|400px|center|]] | [[Image:Nrf2gene.jpg|400px|center|]] | ||
Revision as of 03:06, 13 December 2021
Keap1-Nrf2 Complex
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Lo, S.-C., Li, X., Henzl, M.T., Beamer, L.J. and Hannink, M. (2006), Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. The EMBO Journal, 25: 3605-3617. https://doi.org/10.1038/sj.emboj.7601243
- ↑ Pitoniak, A., & Bohmann, D. (2015). Mechanisms and functions of Nrf2 signaling in Drosophila. Free radical biology & medicine, 88(Pt B), 302–313. https://doi.org/10.1016/j.freeradbiomed.2015.06.020
- ↑ Data available in https://www.ncbi.nlm.nih.gov/gene/9817
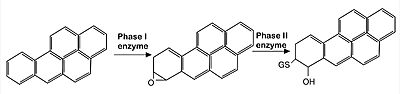
- ↑ Mathers J, Fraser JA, McMahon M, Saunders RD, Hayes JD, McLellan LI. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004;(71):157-76. doi: 10.1042/bss0710157. PMID: 15777020.
- ↑ Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001 Jun 2;477(1-2):7-21. doi: 10.1016/s0027-5107(01)00091-4. PMID: 11376682.
- ↑ Ceconi C, Boraso A, Cargnoni A, Ferrari R. Oxidative stress in cardiovascular disease: myth or fact? Arch Biochem Biophys. 2003 Dec 15;420(2):217-21. doi: 10.1016/j.abb.2003.06.002. PMID: 14654060.
- ↑ Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003 Nov 28;278(48):48021-9. doi: 10.1074/jbc.M308439200. Epub 2003 Sep 10. PMID: 12968018.
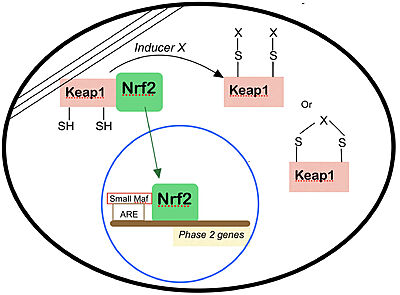
- ↑ 8.0 8.1 Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11908-13. doi: 10.1073/pnas.172398899. Epub 2002 Aug 22. PMID: 12193649; PMCID: PMC129367.
- ↑ 9.0 9.1 9.2 Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004 Nov;10(11):549-57. doi: 10.1016/j.molmed.2004.09.003. PMID: 15519281.
- ↑ Sasaki, H. et al. (2002) Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 277, 44765–44771
- ↑ Chan, K. and Kan, Y.W. (1999) Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U. S. A. 96, 12731–12736
- ↑ Enomoto, A. et al. (2001) High sensitivity of Nrf2 knockout mice to Review TRENDS in Molecular Medicine Vol.10 No.11 November 2004 555 www.sciencedirect.com acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59, 169–177
- ↑ Goldring, C.E. et al. (2004) Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology 39, 1267–1276
- ↑ Ramos-Gomez, M. et al. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 98, 3410–3415
- ↑ Cho, H-Y. et al. (2002) Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26, 175–182