User:Mathias Bortoletto Dunker/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 54: | Line 54: | ||
98, 3410–3415</ref> | 98, 3410–3415</ref> | ||
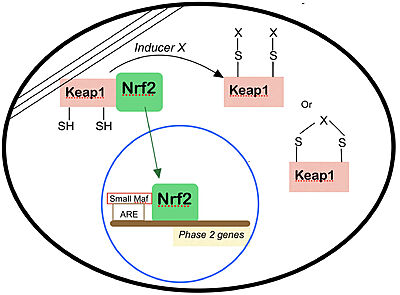
| - | According to Motohashi and Yamamoto,<ref name="super"/> Nrf2 is also an important regulator of oxidative-stress inducible genes, including heme oxygenase-1 and peroxiredoxin 1. In a 2002 study<ref> Cho, H-Y. et al. (2002) Role of NRF2 in protection against hyperoxic | + | According to Motohashi and Yamamoto,<ref name="super"/> Nrf2 is also an important regulator of oxidative-stress inducible genes, including heme oxygenase-1 and peroxiredoxin 1. In a 2002 study<ref> Cho, H-Y. et al. (2002) Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26, 175–182</ref>, a single-nucleotide polymorphism was detected in the promoter region of the Nrf2 gene of the mouse strain C57BL/6J, which is sensitive to hyperoxic stress. Supporting this, Nrf2-null mutant mice were found to be highly susceptible to hyperoxic lung injury. The impaired defense mechanisms against oxidative stress that are observed in the nrf2-null mutant mice could have resulted from the accumulation of reactive oxygen species (ROS) in the absence of Nrf2. A combination of electron paramagnetic resonance (EPR) and spin-probe kinetic analysis confirmed that there is a substantial decrease in the ability of nrf2-null mutant liver and kidney to eliminate ROS. This impaired elimination of ROS was exacerbated in aging female animals. Consistent with this result, old female Nrf2-deficient mice with an ICR genetic background often developed severe lupus-like autoimmune nephritis. Because ROS have a prominent role in the pathogenesis of nephritis, the accumulation of ROS due to Nrf2 deficiency must have exacerbated the mild glomerular lesions that are inherent to the ICR strain of mice. |
| - | lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26, 175–182</ref>, a single-nucleotide polymorphism was detected in the promoter region of the Nrf2 gene of the mouse strain C57BL/6J, which is sensitive to hyperoxic stress. Supporting this, Nrf2-null mutant mice were found to be highly susceptible to hyperoxic lung injury. The impaired defense mechanisms against oxidative stress that are observed in the nrf2-null mutant mice could have resulted from the accumulation of reactive oxygen species (ROS) in the absence of Nrf2. A combination of electron paramagnetic resonance (EPR) and spin-probe kinetic analysis confirmed that there is a substantial decrease in the ability of nrf2-null mutant liver and kidney to eliminate ROS. This impaired elimination of ROS was exacerbated in aging female animals. Consistent with this result, old female Nrf2-deficient mice with an ICR genetic background often developed severe lupus-like autoimmune nephritis. Because ROS have a prominent role in the pathogenesis of nephritis, the accumulation of ROS due to Nrf2 deficiency must have exacerbated the mild glomerular lesions that are inherent to the ICR strain of mice. | + | |
[[Image:Met.jpg|400px|center|]] | [[Image:Met.jpg|400px|center|]] | ||
| Line 61: | Line 60: | ||
== Structural and Genetic information of Nrf2 == | == Structural and Genetic information of Nrf2 == | ||
| - | NRF2 possesses <scene name='89/898348/Nrf2inteirafinal/1'>seven highly conserved domains</scene> called NRF2-ECH homology (Neh) domains. Their names and function is as follows: | + | NRF2 is 605 amino acids long and possesses <scene name='89/898348/Nrf2inteirafinal/1'>seven highly conserved domains</scene> called NRF2-ECH homology (Neh) domains <ref>Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034</ref>. Their names and function is as follows: |
<scene name='89/898348/Nrf2dom1/1'>Neh1 domain</scene> (orange): A CNC-bZIP domain that allows Nrf2 to heterodimerize with small Maf proteins. | <scene name='89/898348/Nrf2dom1/1'>Neh1 domain</scene> (orange): A CNC-bZIP domain that allows Nrf2 to heterodimerize with small Maf proteins. | ||
| - | <scene name='89/898348/Nrf2dom2/1'>Neh2 domain</scene> (dark blue): allows for <scene name='89/898348/Neh2ultima/1'>binding of NRF2 to Keap1.</scene> | + | <scene name='89/898348/Nrf2dom2/1'>Neh2 domain</scene> (dark blue): allows for <scene name='89/898348/Neh2ultima/1'>binding of NRF2 to Keap1, through teo dagrons called DLG and ETGE motifs. </scene> |
<scene name='89/898348/Nrf2dom3/1'>Neh3</scene>, <scene name='89/898348/Nrf2dom4/1'>Neh4</scene> and <scene name='89/898348/Nrf2dom5/1'>Neh5</scene> domain (red, blue and light blue): may play a role in NRF2 protein stability and may act as a transactivation domain, interacting with component of the transcriptional apparatus (Neh3) or to a protein called cAMP Response Element Binding Protein (Neh4 and Neh5), which possesses intrinsic histone acetyltransferase activity. | <scene name='89/898348/Nrf2dom3/1'>Neh3</scene>, <scene name='89/898348/Nrf2dom4/1'>Neh4</scene> and <scene name='89/898348/Nrf2dom5/1'>Neh5</scene> domain (red, blue and light blue): may play a role in NRF2 protein stability and may act as a transactivation domain, interacting with component of the transcriptional apparatus (Neh3) or to a protein called cAMP Response Element Binding Protein (Neh4 and Neh5), which possesses intrinsic histone acetyltransferase activity. | ||
| - | <scene name='89/898348/Nrf2dom6/2'>Neh6 domain</scene>(yellow): may contain a degron that is involved in a redox-insensitive process of degradation of NRF2. This occurs even in stressed cells, which normally extend the half-life of NRF2 protein relative to unstressed conditions by suppressing other degradation pathways. | + | <scene name='89/898348/Nrf2dom6/2'>Neh6 domain</scene>(yellow): may contain a degron that is not recognized by Keap1, but it is instead involved in a redox-insensitive process of E3 ubiquitin ligase β-TrCP degradation of NRF2. This occurs even in stressed cells, which normally extend the half-life of NRF2 protein relative to unstressed conditions by suppressing other degradation pathways. |
| + | |||
| + | <scene name='89/898348/Nrf2dom7/1'>Nh7 domain </scene>(green): interacts with the DNA-binding domain of retinoic X receptor alfa. | ||
| + | |||
| - | <scene name='89/898348/Nrf2dom7/1'>Nh7 domain </scene>(green): | ||
The gene responsible for Nrf2 coding in humans is called HAGRID 283, and its sequence of bases is available in the [https://genomics.senescence.info/genes/entry.php?hgnc=NFE2L2 Human Ageing Genomic Resources] database, and it is as follows: | The gene responsible for Nrf2 coding in humans is called HAGRID 283, and its sequence of bases is available in the [https://genomics.senescence.info/genes/entry.php?hgnc=NFE2L2 Human Ageing Genomic Resources] database, and it is as follows: | ||
[[Image:Nrf2gene.jpg|400px|center|]] | [[Image:Nrf2gene.jpg|400px|center|]] | ||
Revision as of 03:21, 13 December 2021
Keap1-Nrf2 Complex
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Lo, S.-C., Li, X., Henzl, M.T., Beamer, L.J. and Hannink, M. (2006), Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. The EMBO Journal, 25: 3605-3617. https://doi.org/10.1038/sj.emboj.7601243
- ↑ Pitoniak, A., & Bohmann, D. (2015). Mechanisms and functions of Nrf2 signaling in Drosophila. Free radical biology & medicine, 88(Pt B), 302–313. https://doi.org/10.1016/j.freeradbiomed.2015.06.020
- ↑ Data available in https://www.ncbi.nlm.nih.gov/gene/9817
- ↑ Mathers J, Fraser JA, McMahon M, Saunders RD, Hayes JD, McLellan LI. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004;(71):157-76. doi: 10.1042/bss0710157. PMID: 15777020.
- ↑ Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001 Jun 2;477(1-2):7-21. doi: 10.1016/s0027-5107(01)00091-4. PMID: 11376682.
- ↑ Ceconi C, Boraso A, Cargnoni A, Ferrari R. Oxidative stress in cardiovascular disease: myth or fact? Arch Biochem Biophys. 2003 Dec 15;420(2):217-21. doi: 10.1016/j.abb.2003.06.002. PMID: 14654060.
- ↑ Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003 Nov 28;278(48):48021-9. doi: 10.1074/jbc.M308439200. Epub 2003 Sep 10. PMID: 12968018.
- ↑ 8.0 8.1 Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11908-13. doi: 10.1073/pnas.172398899. Epub 2002 Aug 22. PMID: 12193649; PMCID: PMC129367.
- ↑ 9.0 9.1 9.2 Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004 Nov;10(11):549-57. doi: 10.1016/j.molmed.2004.09.003. PMID: 15519281.
- ↑ Sasaki, H. et al. (2002) Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 277, 44765–44771
- ↑ Chan, K. and Kan, Y.W. (1999) Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U. S. A. 96, 12731–12736
- ↑ Enomoto, A. et al. (2001) High sensitivity of Nrf2 knockout mice to Review TRENDS in Molecular Medicine Vol.10 No.11 November 2004 555 www.sciencedirect.com acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59, 169–177
- ↑ Goldring, C.E. et al. (2004) Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology 39, 1267–1276
- ↑ Ramos-Gomez, M. et al. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 98, 3410–3415
- ↑ Cho, H-Y. et al. (2002) Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26, 175–182
- ↑ Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034