User:Mathias Bortoletto Dunker/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 83: | Line 83: | ||
== Structural and Genetic Information of Keap1 == | == Structural and Genetic Information of Keap1 == | ||
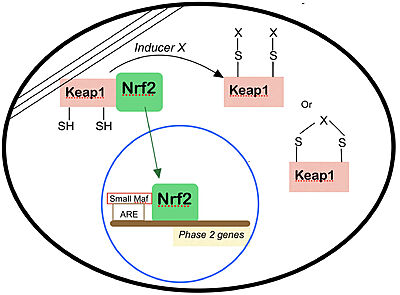
| - | Keap1 belongs to the metazoan superfamily of BTB-[https://en.wikipedia.org/wiki/Kelch_protein Kelch] proteins, a widespread group of proteins that contain multiple Kelch motifs. The Kelch domain generally occurs as a set of five to seven kelch tandem repeats that form a <scene name='89/898348/Betaprop/1'>β-propeller tertiary structure.</scene>. In mice, Keap1 has 624 amino acids, composing five different domains with the following names and caracteristics: | + | Keap1 belongs to the metazoan superfamily of BTB-[https://en.wikipedia.org/wiki/Kelch_protein Kelch] proteins, a widespread group of proteins that contain multiple Kelch motifs. The Kelch domain generally occurs as a set of five to seven kelch tandem repeats that form a <scene name='89/898348/Betaprop/1'>β-propeller tertiary structure.</scene>. In <scene name='89/898348/Keap1mice/1'>mice</scene>, Keap1 has 624 amino acids, composing five different domains with the following names and caracteristics: |
NTR: N-terminal region. | NTR: N-terminal region. | ||
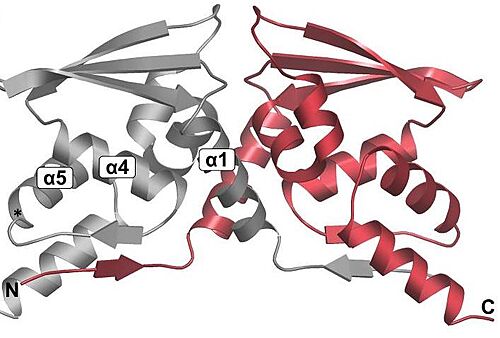
| - | BTB: The BTB/POZ domain (bric-a-brac, tramtrack, broad complex/Poxvirus zinc finger) is an evolutionary conserved domain also found in actin-binding proteins, zinc finger transcription factors, and substrate specific adaptor proteins in Cullin3 (Cul3)-based E3 ubiquitin ligase complexes <ref name="res" />. In many cases, this is a protein-protein interaction domain that mediates dimerization, a process that is required for the binding with Nrf2. | + | <scene name='89/898348/Btb/1'>BTB</scene>: The BTB/POZ domain (bric-a-brac, tramtrack, broad complex/Poxvirus zinc finger) is an evolutionary conserved domain also found in actin-binding proteins, zinc finger transcription factors, and substrate specific adaptor proteins in Cullin3 (Cul3)-based E3 ubiquitin ligase complexes <ref name="res" />. In many cases, this is a protein-protein interaction domain that mediates dimerization, a process that is required for the binding with Nrf2. |
[[Image:DimmerBTB.jpg|500px|center|]] | [[Image:DimmerBTB.jpg|500px|center|]] | ||
Overall fold of the Keap1 BTB crystallographic dimer as a cartoon representation. The N and C-termini, and key alpha-helical secondary structural elements are labelled for one BTB monomer.<ref>Cleasby A, Yon J, Day PJ, et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9(6):e98896. Published 2014 Jun 4. doi:10.1371/journal.pone.0098896</ref>. | Overall fold of the Keap1 BTB crystallographic dimer as a cartoon representation. The N and C-termini, and key alpha-helical secondary structural elements are labelled for one BTB monomer.<ref>Cleasby A, Yon J, Day PJ, et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9(6):e98896. Published 2014 Jun 4. doi:10.1371/journal.pone.0098896</ref>. | ||
| + | |||
IVR: | IVR: | ||
| - | DGR (Kelch): | + | DGR (Kelch):The Kelch domain is a monomer and is comprised of six Kelch repeats that form a symmetric, six-bladed â-propeller structure. The structure reveals that the Kelch repeat motif is defined by highly conserved glycine, tyrosine, and tryptophan residues. There are eight cysteine residues, none of them engaged in disulfide bonds. Interestingly, the first blade of the propeller consists of three strands from the N terminus of the protein and one strand from the C terminus, thus bringing the carboxy terminal region of Keap1 into close proximity to the IVR. Even thought the Kelch domain is not responsible for sensing the introduction of oxidative substances (that is the role of Cys273 and Cys288 in the IVR domain), it is still a highly reactive site and it is through the Kelch domain that the Keap1 protein is able to bind itself to the Neh2 domain of Nrf2. <scene name='89/898348/Flu2/1'>β-propeller tertiary structure.</scene> |
CTR: | CTR: | ||
Revision as of 06:54, 13 December 2021
Keap1-Nrf2 Complex
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Lo, S.-C., Li, X., Henzl, M.T., Beamer, L.J. and Hannink, M. (2006), Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. The EMBO Journal, 25: 3605-3617. https://doi.org/10.1038/sj.emboj.7601243
- ↑ Pitoniak, A., & Bohmann, D. (2015). Mechanisms and functions of Nrf2 signaling in Drosophila. Free radical biology & medicine, 88(Pt B), 302–313. https://doi.org/10.1016/j.freeradbiomed.2015.06.020
- ↑ Data available in https://www.ncbi.nlm.nih.gov/gene/9817
- ↑ Mathers J, Fraser JA, McMahon M, Saunders RD, Hayes JD, McLellan LI. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004;(71):157-76. doi: 10.1042/bss0710157. PMID: 15777020.
- ↑ Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001 Jun 2;477(1-2):7-21. doi: 10.1016/s0027-5107(01)00091-4. PMID: 11376682.
- ↑ Ceconi C, Boraso A, Cargnoni A, Ferrari R. Oxidative stress in cardiovascular disease: myth or fact? Arch Biochem Biophys. 2003 Dec 15;420(2):217-21. doi: 10.1016/j.abb.2003.06.002. PMID: 14654060.
- ↑ Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003 Nov 28;278(48):48021-9. doi: 10.1074/jbc.M308439200. Epub 2003 Sep 10. PMID: 12968018.
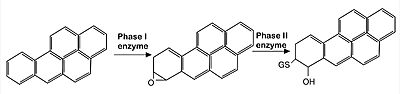
- ↑ 8.0 8.1 Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11908-13. doi: 10.1073/pnas.172398899. Epub 2002 Aug 22. PMID: 12193649; PMCID: PMC129367.
- ↑ 9.0 9.1 Albena T. Dinkova-Kostova, W. David Holtzclaw, and Thomas W. Kensler. The Role of Keap1 in Cellular Protective Responses. American Chemical Society, August 2, 2005.
- ↑ Nobunao Wakabayashi, Albena T. Dinkova-Kostova, W. David Holtzclaw, Moon-Il Kang, Akira Kobayashi, Masayuki Yamamoto, Thomas W. Kensler, and Paul Talalay. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. PNAS February 17, 2004 vol. 101 no. 7.
- ↑ 11.0 11.1 11.2 Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004 Nov;10(11):549-57. doi: 10.1016/j.molmed.2004.09.003. PMID: 15519281.
- ↑ Sasaki, H. et al. (2002) Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 277, 44765–44771
- ↑ Chan, K. and Kan, Y.W. (1999) Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U. S. A. 96, 12731–12736
- ↑ Enomoto, A. et al. (2001) High sensitivity of Nrf2 knockout mice to Review TRENDS in Molecular Medicine Vol.10 No.11 November 2004 555 www.sciencedirect.com acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59, 169–177
- ↑ Goldring, C.E. et al. (2004) Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology 39, 1267–1276
- ↑ Ramos-Gomez, M. et al. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 98, 3410–3415
- ↑ Cho, H-Y. et al. (2002) Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26, 175–182
- ↑ Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034
- ↑ https://www.annualreviews.org/doi/full/10.1146/annurev-cancerbio-030518-055627, CC BY 4.0, https://commons.wikimedia.org/w/index.php?curid=94063579
- ↑ Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034
- ↑ Cleasby A, Yon J, Day PJ, et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9(6):e98896. Published 2014 Jun 4. doi:10.1371/journal.pone.0098896