Dihydrofolate reductase
From Proteopedia
| Line 42: | Line 42: | ||
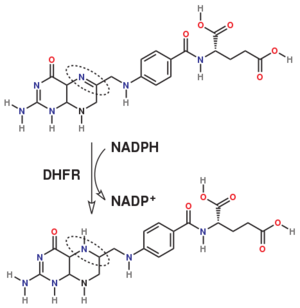
DHFR is thought to proceed in a multi-step mechanism. Once NADPH and DHF are bound to the E. coli enzyme, DHF is first protonated and then reduced through a hydride transfer from NADPH.<ref>doi:10.1073/pnas.1415940111</ref> Substrate binding and product release is thought to have a definite choreography, with fresh NADPH binding before THF is released. | DHFR is thought to proceed in a multi-step mechanism. Once NADPH and DHF are bound to the E. coli enzyme, DHF is first protonated and then reduced through a hydride transfer from NADPH.<ref>doi:10.1073/pnas.1415940111</ref> Substrate binding and product release is thought to have a definite choreography, with fresh NADPH binding before THF is released. | ||
| - | + | {{Template:Button Toggle Animation2}} | |
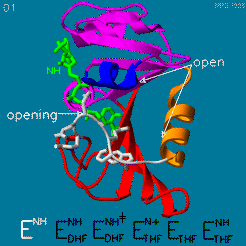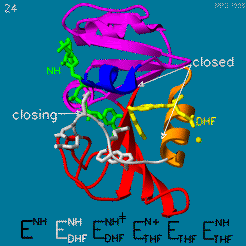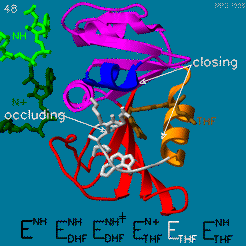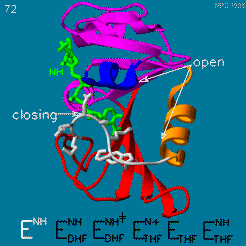
| - | The | + | The <scene name='82/82636/Substrate_product/3'>domain orientation and the Met20 loop conformations</scene> changes during the catalytic cycle. A model by M. R. Sawaya and J. Kraut from 1997 based on six isomorphous crystal structures shows the envisioned sequence of events in the movie below <ref>doi:10.1021/bi962337c</ref>. The original web page of this movie is available on the [https://web.archive.org/web/19991003061611/http://chem-faculty.ucsd.edu/kraut/dhfr.html web archive]. |
| - | + | [[Image:Dhfr.movie2.gif]] | |
| + | The <scene name='82/82636/Active_site/1'>protonation step</scene> was studied using neutron diffraction and high-resolution X-ray diffractionto visualize the hydrogen atoms that are usually not visible in crystal structures.<ref>doi:10.1073/pnas.1415856111</ref> | ||
| - | + | The hydride transfer is thought to involve hydride tunneling, supported by temperature-dependent kinetic isotope effects. Tunneling is a quantum phenomenon explaining how a small particle can cross an activation barrier even when it lacks sufficient activation energy. <ref>doi:10.3390/quantum3010006</ref> | |
| - | + | ||
=== Inhibitors === | === Inhibitors === | ||
Revision as of 16:54, 7 January 2022
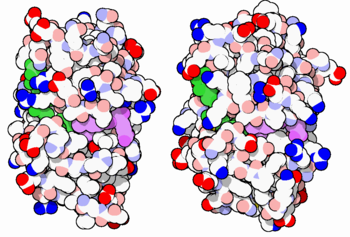
The enzyme dihydrofolate reductase (DHFR) occurs in all organisms and has been particularly well-studied in the bacterium Escherichia coli and in humans[1][2][3]. It catalyzes the reduction of dihydrofolate to tetrahydrofolate, with NADPH acting as hydride donor. The human enzyme is a target for developing inhibitors used in anti-cancer chemotherapies[4], while the bacterial enzymes are targets for developing inhibitors as antibiotics. DHFR is a model enzyme for studying the kinetics, mechanism, and inhibition of enzymatic reactions and the underlying structure and conformational dynamics.
Contents |
DHFR from different organisms
DHFR is found in all organisms. Some bacteria acquire resistance to DHFR inhibitors through expressing a second form of DHFR coded on a plasmid. The enzymes from E. coli (ecDHFR) and humans (hDHFR) have similar folds, while the plasmid-encoded enzyme has an unrelated fold. In humans, DHFR is expressed in most tissues[1], and there are two genes, DHFR and DHFR2/DHFRL1, the latter targeted to mitochondria[5]. Mice and rats lack the second gene but also show DHFR activity in mitochondria[6].
Reactions catalyzed
Dihydrofolate reductase (DHFR, 1.5.1.3 [2]) is an enzyme which uses the co-factor NADPH as electron donor. It catalyzes the reduction of as NADPH is oxidized to NADP+. The mammalian enzymes also accept folic acid as a substrate, reducing it to THF. This allows the use of folic acid, which is easier to synthesize than DHF or THF, to fortify food.[7]. Some bacterial enzymes also accept folic acid as a substrate [8] but it acts as a competitive inhibitor in the E. coli enzyme.
The folate is a form of the essential vitamin B9. Folate is not part of our natural diet (it contains dihydrofolate and tetrahydrofolate, sometimes as a poly-glutamate conjugate) but is bioavailable and simpler to synthesize.
Relevance
DHFR forms a complex with thymidylate synthase (TS)[9]. Both enzymes participate in the biosynthesis of pyrimidine.[10]
| |||||||||||
See also
3D Structures of Dihydrofolate reductase
Dihydrofolate reductase 3D structures
Additional Resources
- For additional information, see: Cancer.
- See also Molecular Playground/DHFR.
References
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Karsten Theis, Alexander Berchansky, Joel L. Sussman, Tzvia Selzer, Jaime Prilusky, Eric Martz, Eran Hodis, David Canner