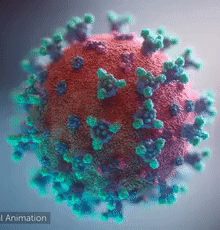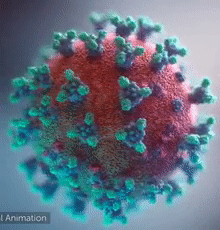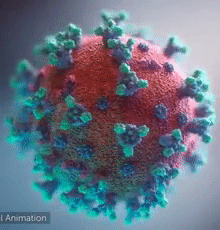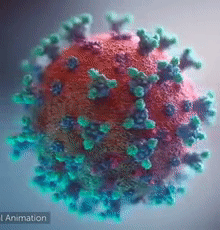
Coronavirus Disease 2019 (COVID-19)
From Proteopedia
(Difference between revisions)
| Line 47: | Line 47: | ||
* 31-Mar-2020 ''NY Times'' [http://www.nytimes.com/2020/03/31/health/cdc-masks-coronavirus.html C.D.C. Weighs Advising Everyone to Wear a Mask]. | * 31-Mar-2020 ''NY Times'' [http://www.nytimes.com/2020/03/31/health/cdc-masks-coronavirus.html C.D.C. Weighs Advising Everyone to Wear a Mask]. | ||
| - | |||
| - | == 3D structural studies on SARS-CoV-2 virus== | ||
| - | {{Template:COVID Validation}} | ||
| - | |||
| - | * A team of UK & Israeli scientists determined the 3D structures of [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem/Downloads.html '''over 90 fragments, 66 of which are in the active site'']. All the experimental details and results are available [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem.html '''online at the Diamond Light Source''']. | ||
| - | |||
| - | * A team of Chinese scientists determined, by Cryo-EM, the''' coronavirus spike receptor-binding domain complexed with its receptor ACE2 PDB-ID''' [[6lzg]] (To be published). | ||
| - | |||
| - | * A team of US and Chinese scientists determined the crystal structure of '''2019-nCoV spike receptor-binding domain bound with ACE2''' [[6m0j]] | ||
| - | |||
| - | * A team of US scientists determined, by Cryo-EM, the '''structure of the SARS-CoV-2 spike glycoprotein (open & closed states)''' <ref>PMID:32155444</ref>, PDB-ID [[6vxx]] and [[6vyb]]. See an [[SARS-CoV-2 protein S priming by furin|animation of this transition]]. | ||
| - | |||
| - | * Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody CR3022 [[6w41]] | ||
| - | |||
| - | * Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2 [[6w61]] (To be published). | ||
| - | |||
| - | * Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP [[6w6y]] (To be published). | ||
| - | |||
| - | * Structure of NSP10 - NSP16 Complex from SARS-CoV-2 [[6w75]] (To be published). | ||
| - | |||
| - | * Crystal structure of SARS-CoV-2 nucleocapsid protein N-terminal RNA binding domain [[6m3m]] (To be published). | ||
| - | |||
| - | * Crystal structure of Nsp9 RNA binding protein of SARS CoV-2 [[6w4b]] (To be published). | ||
| - | |||
| - | * Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 [[6w4h]] (To be published). | ||
| - | |||
| - | * A Cryo-EM study by Zhou & colleagues on the structural basis for the '''recognition of the SARS-CoV-2 by full-length human ACE2''' gives insights to the molecular basis for coronavirus recognition and infection<ref>PMID:32132184</ref> [[6m17]]. | ||
| - | |||
| - | * Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 [[6vyo]] (To be published). | ||
| - | |||
| - | * Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate [[6w01]] (To be published). | ||
| - | |||
| - | * Crystal structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose [[6w02]] (To be published). | ||
| - | |||
| - | * Crystal structure of 2019-nCoV chimeric receptor-binding domain complexed with its receptor human ACE2 [[6vw1]] | ||
| - | |||
| - | * Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 [[6vww]] (To be published). | ||
| - | |||
| - | * Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 [[6vxs]] (To be published). | ||
| - | |||
| - | * Crystal structure of the 2019-nCoV HR2 Domain [[6lvn]] (To be published). | ||
| - | |||
| - | * Crystal structure of post fusion core of 2019-nCoV S2 subunit [[6lxt]]. | ||
| - | |||
| - | * Crystal structure of '''SARS-CoV-2 main protease''' provides a basis for design of improved α-ketoamide inhibitors, from the Hilgenfeld lab<ref>PMID:32198291</ref>, Apo Struture: PDB-ID [[6y2e]], and complexes with inhibitors: PDB-ID [[6y2f ]] and [[6y2g]]. | ||
| - | |||
| - | * 3D Structure of '''RNA-dependent RNA polymerase from COVID-19''', a '''major antiviral drug target''' from the Rao lab in Beijing<ref> Gao, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.16.993386 http://doi.org/10.1101/2020.03.16.993386]</ref>. | ||
| - | |||
| - | * Crystal structure of the '''Mpro from COVID-19''' and '''discovery of inhibitors''' in a study by scientists from Shanghai & Beijing<ref>PMID: 32272481</ref>, PDB-ID [[6lu7]]. | ||
| - | |||
| - | * Crystal structure of '''Nsp15 endoribonuclease NendoU from SARS-CoV-2''' in a study by scientists from USA<ref> Kim, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.02.968388 http://doi.org/10.1101/2020.03.02.968388]</ref>, PDB-ID [[6w01]]. | ||
| - | |||
| - | * '''The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics'''. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis<ref name="McLellan" />, PDB-ID [[6vsb]]. | ||
| - | |||
| - | * [http://github.com/thorn-lab/coronavirus_structural_task_force Public resource for the structures from beta-coronavirus with a focus on SARS-CoV and SARS-CoV-2] | ||
== SARS-CoV-2 virus proteins == | == SARS-CoV-2 virus proteins == | ||
Revision as of 11:15, 7 February 2022
| |||||||||||
References
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Jaime Prilusky, Eric Martz, Jurgen Bosch, David Sehnal, Gianluca Santoni