3D structural studies on SARS-CoV-2
From Proteopedia
(Difference between revisions)
(New page: ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> This is a default text for you...) |
|||
| Line 1: | Line 1: | ||
| - | + | [[Image:Ezgif.com-video-to-gif 240px 65pc speed.gif|right|240px|thumb|<span style="font-size:100%">SARS-CoV-2 virus. The <span style="color:blue">spikes</span>, that adorn the <span style="color:red">'''virus surface'''</span>, impart a '''''corona''''' like appearance [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers (Fusion Animation)].</span>]] | |
| - | < | + | <span style="font-size:100%"> |
| - | + | ||
| - | + | ||
| - | == | + | {{Template:COVID Validation}} |
| + | The genome of the SARS-CoV-2 virus codes for 28 proteins: | ||
| + | Out of those, 19 have already been characterized structurally. For the rest there are accurate [[AlphaFold]]2 predicted structures. | ||
| + | <br> | ||
| + | Details of the '''3D structure & function''' of the key proteins & RNA inside the virus can be seen in the ''NY Times''<ref name="NY Times COVID-19">[https://www.nytimes.com/interactive/2020/04/03/science/coronavirus-genome-bad-news-wrapped-in-protein.html ''NY Times'' (3-Apr-2020) Bad News Wrapped in Protein: Inside the Coronavirus Genome]</ref>. | ||
| + | "The first viral protein created inside the infected cell, ORF1ab, is actually a chain of 16 proteins joined together. Two of these proteins act like scissors, snipping the links between the different proteins and freeing them to do their jobs."<ref name="NY Times COVID-19"/> | ||
| - | = | + | [[Image:SARS-CoV-2 Prot Organization.jpg|500px|center|thumb|SARS-CoV-2 Protein Organization, redrawn from NY Times<ref name="NY Times COVID-19"/>]]. |
| - | + | * [[SARS-CoV-2 Coronavirus Main Protease| Main protease]]: it is a cysteine protease that is essential for the viral life cycle. | |
| - | + | * [[SARS-CoV-2_protein_NSP1|NSP1]]: ''Host translation inhibitor nsp1''. Inhibits host translation by interacting with the 40S ribosomal subunit. | |
| - | + | * [[SARS-CoV-2_protein_NSP2|NSP2]]: ''Non-structural protein 2''. | |
| - | + | * [[SARS-CoV-2 enzyme Papain-like|Papain-like proteinase]] | |
| - | + | * [[SARS-CoV-2_protein_NSP4|NSP4]]: ''Non-structural protein 4''. | |
| - | + | * [[SARS-CoV-2 enzyme 3CL-PRO|Proteinase 3CL-PRO]] | |
| - | + | * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2_protein_NSP6|NSP6]]: ''Non-structural protein 6''. | |
| + | * [[SARS-CoV-2_protein_NSP7|NSP7]]: ''Non-structural protein 7''. | ||
| + | * [[SARS-CoV-2_protein_NSP8|NSP8]]: ''Non-structural protein 8''. | ||
| + | * [[SARS-CoV-2_protein_NSP9|NSP9]]: ''Non-structural protein 9''. | ||
| + | * [[SARS-CoV-2_protein_NSP10|NSP10]]: ''Non-structural protein 10''. | ||
| + | * [[SARS-CoV-2 enzyme RdRp|RNA-directed RNA polymerase (RdRp)]] | ||
| + | * [[SARS-CoV-2 enzyme Hel|Helicase (Hel)]]: Multi-functional protein with a zinc-binding domain in N-terminus | ||
| + | * [[SARS-CoV-2 enzyme ExoN|Guanine-N7 methyltransferase (ExoN)]] | ||
| + | * [[SARS-CoV-2 enzyme NendoU|Uridylate-specific endoribonuclease (NendoU)]] | ||
| + | * [[SARS-CoV-2 enzyme 2'-O-MT|2'-O-methyltransferase (2'-O-MT)]]: Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs. | ||
| + | * [[SARS-CoV-2_protein_S|Surface glycoprotein (S)]] and [[SARS-CoV-2 protein S priming by furin]] | ||
| + | * [[SARS-CoV-2 protein ORF3a|ORF3a]]: Forms homotetrameric potassium sensitive ion channels (viroporin) and may modulate virus release. | ||
| + | * [[SARS-CoV-2_protein_E|Protein E]]: Plays a central role in virus morphogenesis and assembly. | ||
| + | * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2_protein_M|Protein M]]: Component of the viral envelope. | ||
| + | * [[SARS-CoV-2 protein ORF6|ORF6]]: Could be a determinant of virus virulence. | ||
| + | * [[SARS-CoV-2 protein ORF7a|ORF7a]]: Non-structural protein which is dispensable for virus replication in cell culture. | ||
| + | * [[SARS-CoV-2 protein ORF8|ORF8]]: Open Reading Frame 8. | ||
| + | * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2_protein_N|Protein N]]: Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP). | ||
| + | * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2 protein ORF10|ORF10]]: It is currently unclear whether this region translates into a functional protein. | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 11:14, 7 February 2022
SARS-CoV-2 virus. The spikes, that adorn the virus surface, impart a corona like appearance (Fusion Animation).
| The quality of SARS-CoV-2 experimentally determined structures varies widely (Grabowski et al., 2021). Validated and corrected structures can be obtained from COVID19.BioReproducibility.Org. |
The genome of the SARS-CoV-2 virus codes for 28 proteins:
Out of those, 19 have already been characterized structurally. For the rest there are accurate AlphaFold2 predicted structures.
Details of the 3D structure & function of the key proteins & RNA inside the virus can be seen in the NY Times[1].
"The first viral protein created inside the infected cell, ORF1ab, is actually a chain of 16 proteins joined together. Two of these proteins act like scissors, snipping the links between the different proteins and freeing them to do their jobs."[1]
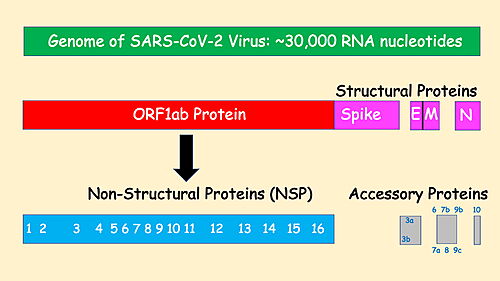
SARS-CoV-2 Protein Organization, redrawn from NY Times[1]
- Main protease: it is a cysteine protease that is essential for the viral life cycle.
- NSP1: Host translation inhibitor nsp1. Inhibits host translation by interacting with the 40S ribosomal subunit.
- NSP2: Non-structural protein 2.
- Papain-like proteinase
- NSP4: Non-structural protein 4.
- Proteinase 3CL-PRO
- AlphaFold2 Theoretical Model NSP6: Non-structural protein 6.
- NSP7: Non-structural protein 7.
- NSP8: Non-structural protein 8.
- NSP9: Non-structural protein 9.
- NSP10: Non-structural protein 10.
- RNA-directed RNA polymerase (RdRp)
- Helicase (Hel): Multi-functional protein with a zinc-binding domain in N-terminus
- Guanine-N7 methyltransferase (ExoN)
- Uridylate-specific endoribonuclease (NendoU)
- 2'-O-methyltransferase (2'-O-MT): Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs.
- Surface glycoprotein (S) and SARS-CoV-2 protein S priming by furin
- ORF3a: Forms homotetrameric potassium sensitive ion channels (viroporin) and may modulate virus release.
- Protein E: Plays a central role in virus morphogenesis and assembly.
- AlphaFold2 Theoretical Model Protein M: Component of the viral envelope.
- ORF6: Could be a determinant of virus virulence.
- ORF7a: Non-structural protein which is dispensable for virus replication in cell culture.
- ORF8: Open Reading Frame 8.
- AlphaFold2 Theoretical Model Protein N: Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP).
- AlphaFold2 Theoretical Model ORF10: It is currently unclear whether this region translates into a functional protein.
