User:George G. Papadeas/Sandbox VKOR
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Introduction== | == Introduction== | ||
=== History of VKOR === | === History of VKOR === | ||
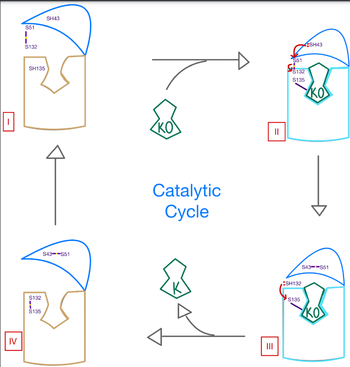
| - | Vitamin K epoxide reductase (VKOR) is an enzyme that, as its name implies, promotes the reduction of vitamin K epoxide (KO). VKOR is a transmembrane protein spanning the endoplasmic reticulum and composed of 4 transmembrane helical proteins. One of its primary roles is to assist in blood coagulation by regenerating hydroquinone (KH2). KH2 acts as a | + | Vitamin K epoxide reductase (VKOR) is an enzyme that, as its name implies, promotes the reduction of vitamin K epoxide (KO). VKOR is a transmembrane protein spanning the endoplasmic reticulum and composed of 4 transmembrane helical proteins. One of its primary roles is to assist in blood coagulation by regenerating hydroquinone (KH2). KH2 acts as a γ-carboxylase cofactor that drives the γ-carboxylation of several coagulation factors. Structural characterization of VKOR has been difficult, though, due to its in vitro instability. Nonetheless, a near perfect atomic structure has been determined utilization anticoagulant stabilization and VKOR-like [http://https://pubmed.ncbi.nlm.nih.gov/33154105/ homologs]. |
=== Function and Biological Role === | === Function and Biological Role === | ||
=== Author's Notes === | === Author's Notes === | ||
| - | As previously mentioned, the VKOR structure has | + | As previously mentioned, the VKOR structure has been challenging to qualify. Thus it is important to note that to date all VKOR structures discovered were done so from 2 methods. First, crystal structures of Human VKOR were captured with a bound substrate (KO) or vitamin K antagonist (VKA). VKA substrates utilized were anticoagulants, namely Warfarin, brodifacoum, phenindione, and chlorophacinone. Second, VKOR-like homologs, specifically isolated from the pufferfish ''Takifugu rubripes'', aided in structure classification as well. |
== Structural Highlights== | == Structural Highlights== | ||
Revision as of 22:40, 26 March 2022
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644