Sandbox Reserved 1700
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
==== Toggle Switch ==== | ==== Toggle Switch ==== | ||
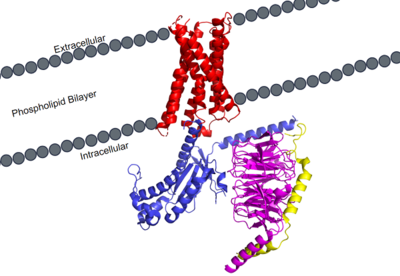
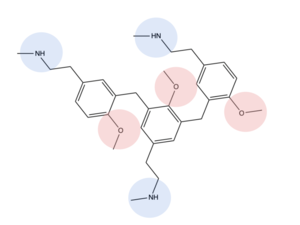
| - | Certain residues that lie in the ligand binding pocket can act as molecular switches to turn the GPCR “on” or “off” and are fittingly called toggle switches. Toggle switches in receptors are essential in interacting with the ligand upon binding, as they can then initiate the transmission of the molecular signal through the protein. Trp-336 has been known to act as this toggle switch in GPCR’s of the A family, however in MRGPRX2, the residue has been replaced to a <scene name='90/904305/Glycine_toggle_switch/7'>glycine</scene>. This is a significant modification to the receptor structure. By replacing the large tryptophan residue with a small glycine, the membrane helicies, especially helix 7 on which the toggle switch is found, can pack closer together. The ligands that interact with MRGPRX2 then | + | Certain residues that lie in the ligand binding pocket can act as molecular switches to turn the GPCR “on” or “off” and are fittingly called toggle switches. Toggle switches in receptors are essential in interacting with the ligand upon binding, as they can then initiate the transmission of the molecular signal through the protein. Trp-336 has been known to act as this toggle switch in GPCR’s of the A family, however in MRGPRX2, the residue has been replaced to a <scene name='90/904305/Glycine_toggle_switch/7'>glycine</scene>. This is a significant modification to the receptor structure. By replacing the large tryptophan residue with a small glycine, the membrane helicies, especially helix 7 on which the toggle switch is found, can pack closer together. The ligands that interact with MRGPRX2 then bind much closer to the surface of the receptor, as opposed to deeper within the helices. This significant change can be visualized in the image above. This significantly changes what structures of ligands are able to interact with this receptor and therefore what types of molecules can activate the Human Itch GPCR. More details about what kinds of ligands bind to this receptor are discussed later. |
==== Sodium Site ==== | ==== Sodium Site ==== | ||
Revision as of 23:45, 28 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
MRGPRX2 Human Itch G-Protein Coupled Receptor (GPCR)
| |||||||||||
References
- ↑ Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842. doi: 10.1038/nrd.2017.178. Epub, 2017 Oct 27. PMID:29075003 doi:http://dx.doi.org/10.1038/nrd.2017.178
- ↑ Basith S, Cui M, Macalino SJY, Park J, Clavio NAB, Kang S, Choi S. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front Pharmacol. 2018 Mar 9;9:128. doi: 10.3389/fphar.2018.00128. eCollection, 2018. PMID:29593527 doi:http://dx.doi.org/10.3389/fphar.2018.00128
- ↑ 3.0 3.1 Cao C, Kang HJ, Singh I, Chen H, Zhang C, Ye W, Hayes BW, Liu J, Gumpper RH, Bender BJ, Slocum ST, Krumm BE, Lansu K, McCorvy JD, Kroeze WK, English JG, DiBerto JF, Olsen RHJ, Huang XP, Zhang S, Liu Y, Kim K, Karpiak J, Jan LY, Abraham SN, Jin J, Shoichet BK, Fay JF, Roth BL. Structure, function and pharmacology of human itch GPCRs. Nature. 2021 Dec;600(7887):170-175. doi: 10.1038/s41586-021-04126-6. Epub 2021, Nov 17. PMID:34789874 doi:http://dx.doi.org/10.1038/s41586-021-04126-6
- ↑ 4.0 4.1 Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP. Structure, function and pharmacology of human itch receptor complexes. Nature. 2021 Dec;600(7887):164-169. doi: 10.1038/s41586-021-04077-y. Epub 2021, Nov 17. PMID:34789875 doi:http://dx.doi.org/10.1038/s41586-021-04077-y