G-Protein Coupled Receptors
G-protein coupled receptors(GCPRs) are a large family of cell surface membrane proteins. Once bound to a wide variety of extracellular ligands, GCPRs undergo a conformational change and relay information to intracellular secondary messengers [1]. This G protein activation results in a cellular response dependent on the ligand bound and location of the GPCR in the body. GCPRs can be broken down into five families: the rhodopsin family (class A, 701 members), the secretin family (class B, 15 members), the adhesion family (24 members), the glutamate family (class C, 15 members), and the frizzled/taste family (class F, 24 members) [2]. All of the families have a similar transmembrane (TM) domain consisting of seven ɑ-helices complexed with intracellular G proteins.
Class A GCPRs
Class A GPCRS or rhodopsin-like GPCRS are the largest and most studied type of GPCRS. Due to the diversity of these receptors they are found in many aspects of physiology. They are characterized by conserved motifs including DRY, PIF, Sodium Binding, and the CWxP [3]. A common example of a class A GPCRS is the β2AR receptor (PDB: 3sn6).
MRGPRs
Structure with
Structure with
The human itch GPCR, or Mas-related G-protein coupled receptor (MRGPR), is a Class A GPCR found in human sensory neurons and is responsible for the sensation of “itching” caused by skin irritation and diseases, insect bites, and hypersensitivity to certain drugs. There are currently four groups consisting of MRGPRX1, MRGPRX2, MRGPRX3, and MRGPRX4. In particular, MRGPRX4 is responsible for cholestatic itch while MRGPRX2 regulates degranulation and hypersensitivity itch reactions [4]. These two, chiefly MRGPRX2, are often targets for drugs that result in mast cell degranulation and hypersensitivity side effects. In comparison to other Class A GPCRs, MRGPRX2 binds to an even wider range of ligands, including agonists such as cations and peptides.
Cation
(R)-ZINC-3573 is a cation ligand that binds to MRGPRX2. Its N-dimethyl is inserted into a cavity of aromatic amino acid residues Phe-170, Trp-243, and Phe-244. In this cavity, it makes ion pairs with Asp-184 and Glu-164. It is then stabilized by stacking its ring with Trp-248 and the Cys-168 to Cys-180 disulfide bond [5].
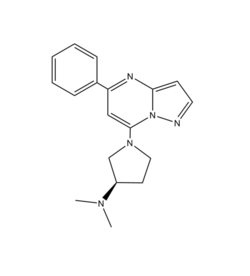
Figure X: Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.
Peptide
Cortistatin-14(structure with cortistatin) is one of the peptide ligands that binds to MRGPRX2. Cortistatin-14 interacts with the binding pocket through an electrostatic (ZIZ) interaction in sub-pocket 1 between Lys-3 on the peptide and Glu-164 and Asp-184 on MRGPRX2. Additionally, there are hydrophobic interactions in sub-pocket 2 between the peptide and the binding pocket due to the large hydrophobic amino acids on Cortistatin-14.
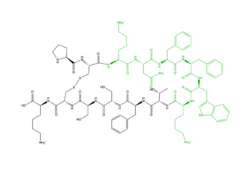
Figure X: Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.
Specific Traits
Disulfide bonds
In common Class A GPCRs the disulfide bond associated with the initiation of signal transduction is located on the extracellular domain of the 7 transmembrane helices. The disulfide bond of β2AR, a well studied Class A GPCR, occurs between (TM3) C106 and (EL) C191. This loop crosses through the middle of the extracellular domain, creating a barrier for bulkier substrates. In MRGPRX2, the disulfide bond bond is located between (TM4) C168 and (TM5) C180. This is a TM to TM disulfide bond as compared to a TM to EL disulfide bond seen in typical Class A GPCRs. This lack of interaction with the extracellular loop seen in MRGPRX2 causes the extracellular loop to flip on top of the TM4 and TM5 resulting in an open space for larger substrates to be able to interact with the receptor.
Toggle Switch
In β2AR, and other Class A GPCRs, there is a what is known as a “toggle switch” Trp-286 which puts a limit on how close the TM helices can get to each other due to tryptophan being a very large, bulky amino acid. This results in a deep binding pocket. In contrast, in MRGPRX2 Trp-286 is replaced by Gly-236 [4] [5]. Glycine is a much smaller amino acid and thus allows the helices to close the base of the binding pocket. This causes MRGPRX2 to have a very shallow binding site and consequently allows an even greater number of ligands to be able to bind.
PIF Motif
Another motif that MRGPRX2 differs from typical Class A GPCRs is the PIF/connector motif, which acts as a microswitch. PIF plays a role in connecting the binding pocket to conformational rearrangements required for receptor activation[6]. This motif is located towards the base of the TM domains. In a Class A GPCR, like β2AR, the PIF motif consists of Phe-211, Ile-121, and Phe-282 on TM domains 5, 3, and 6, respectively. However, for MRGPRX2, this motif is replaced with Met-196 and Leu-194 on TM5, Leu-117 on TM3, and Phe-232 on TM6.
DRY Motif
Sodium Binding
MRGPRX2 Signaling Pathway
Function
Conformational Changes
What It Triggers
Clinical Relevance
Many drugs have been linked to the activation of MRGPRX2 as a side effect that causes the sensation of itchiness. Among these drugs are morphine, codeine, and dextromethorphan. These drugs have a similar structure to that of (R)- ZINC-3573, introducing the idea of a similar binding mechanism.
Due to mutations in key structural features of typical Class A GPCRS, MRGPRX2, it is able to bind to a variety of substrates that then mediate the signaling pathway for the sensation of itching.
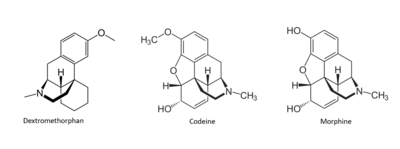
Figure X: Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.
References
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.



