Sandbox Reserved 1700
From Proteopedia
(Difference between revisions)
| Line 84: | Line 84: | ||
==== Disulfide Bonds ==== | ==== Disulfide Bonds ==== | ||
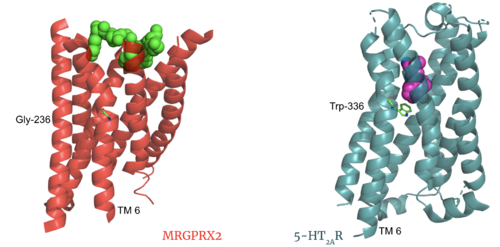
| - | + | The MRGPRX2 disulfide bond is between <scene name='90/904305/Disulfide_bond/1'>Cys-168 and Cys-180</scene> on TM helices 5 and 4, respectively. In other class A GPCRs, this disulfide bond is between and extracellular loop (ECL2) and a TM helix (TM3). For example, the <scene name='90/904306/5ht2a_disulfide/2'>serotonin GPCR</scene> shows this disulfide bond between the ECL2 and TM3. This different disulfide bond location contributes to surface level binding of ligands. | |
| + | [[Image:Screen Shot 2022-03-27 at 5.45.52 PM.png|300px|center|thumb|Overlay of the 5HT2AR and MRGPRX2 TMP for comparison of disulfide bond location.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==== Further Information ==== | ||
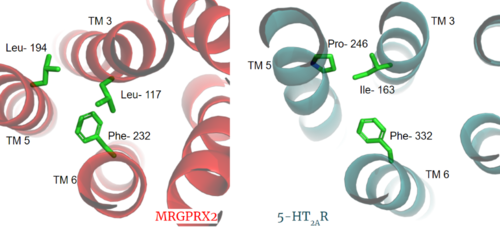
| + | The MRGPRX2 GPCR also contains semi-conserved or fully-conserved motifs that are seen in other class A GPCRs. | ||
| + | |||
| + | =====NPxxY Motif===== | ||
| + | The residues in the <scene name='90/904306/Npxxy_motif/4'>NPxxY Motif</scene> are pivotal for receptor activation in all Class A GPCRs. This motif is conserved in the MRGPRX2 receptor with residues VAL-231, ASP-75, ASN-275, and TYR-279. | ||
| + | |||
| + | =====CWxP Motif===== | ||
| + | The <scene name='90/904306/Cpxw_motif_2/1'>CWxP Motif</scene> is almost fully conserved except for TRP-236, or the toggle switch, which is replaced with GLY-236. CYS-235, LEU-237, and PRO-238 are all conserved. CYS6.47 may play a fundamental role in GPCR activity which would be supported by its conservation in MRGPRX2<ref name="Olivella>PMID:23497259</ref>. | ||
=== Ligands === | === Ligands === | ||
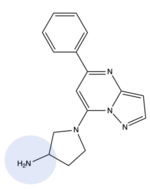
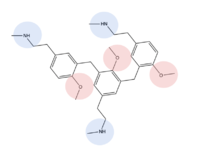
| + | MRGPRX2 binds a wide range of small molecule and peptide ligands. These ligands have positively charged regions which can interact with the negative binding pocket, sub-pocket 1. Some of these larger ligands also have a hydrophobic region which can interact with sub-pocket 2. MRGPRX2 has been known to interact with [https://pubchem.ncbi.nlm.nih.gov/compound/PAMP-12-_human_-porcine#section=Structures PAMP-12], [https://pubchem.ncbi.nlm.nih.gov/compound/44208884 Cortistatin-14], [https://pubchem.ncbi.nlm.nih.gov/compound/118797323 C48/80], and [https://pubchem.ncbi.nlm.nih.gov/compound/95882507 Zinc-3573]<ref name="Yang">PMID: 34789875</ref>,<ref name="Cao">PMID: 34789874</ref>. | ||
| + | [[Image:Image-Screen Shot 2022-03-28 at 8.21.55 PM.png|150px|left|thumb|Zinc-3573]][[Image:80.png|200px|center|thumb|C48/80]] | ||
| + | [[Image:Screen Shot 2022-03-28 at 8.24.59 PM.png|300px|left|thumb|PAMP-12]][[Image:Screen Shot 2022-03-28 at 8.25.58 PM.png|300px|center|thumb|Cortistatin-14]] | ||
| - | [[Image:80.png|300px|left|thumb|C48/80 ligand description]] | ||
| - | == Function == | ||
| + | |||
| + | |||
| + | Ligands are shown with positive regions shown in blue, negative regions in red, and hydrophobic regions in green to demonstrate where they may interact with sub-pockets 1 and 2. <scene name='90/904306/C-14_in_site/4'>Cortistatin-14</scene>, specifically, interacts with sub-pocket 1 through LYS-3 and sub-pocket 2 through LYS-8. | ||
| + | |||
| + | These ligands also share similar structural features with targeted drugs for clinical relevance. MRGPRX2 has been identified as a potential target for drugs such as [https://pubchem.ncbi.nlm.nih.gov/compound/Atracurium#section=2D-Structure Atracurium], [https://pubchem.ncbi.nlm.nih.gov/compound/441290 Rocuronium], [https://pubchem.ncbi.nlm.nih.gov/compound/2764 Ciprofloxacin], and [https://pubchem.ncbi.nlm.nih.gov/compound/149096 Levofloxacin]<ref name="Navines-Ferrer">PMID:30072729</ref>. These drugs have similar structural regions which may interact with sub-pockets 1 and/or 2. | ||
| + | |||
| + | |||
| + | == Function == | ||
| + | GPCRs undergo a transmembrane protein conformational change upon ligand binding. This signal is then transduced to the G-protein allowing for downstream responses. | ||
=== Before Activation === | === Before Activation === | ||
| + | The culmination of different motifs observed in MRGPRX2 compared to other class A GPCRs leads to external membrane ligand binding.The MRGPRX2 GPCR undergoes a much smaller conformational change upon ligand binding compared to other Class A GPCRs due to surface level binding versus deep helix binding. This contrast is represented in the photo below to show the unbound and bound transmembrane proteins of MRGPRX2 and 5-HT2A. | ||
| + | [[Image:Screen Shot 2022-03-282 at 6.59.44 PM.png|500px|center|thumb|Overlay of unbound (transparent) and bound (opaque) transmembrane proteins of both MRGRPX2 (left) and 5-HT2AR (right).]] | ||
=== After Activation === | === After Activation === | ||
| + | After ligand binding and transmembrane protein activation, this signal is sent to the alpha subunit of the G-protein which undergoes its own chemical and conformational changes. The alpha subunit is initially bound with GDP which then is physically replaced by GTP leading to conformational changes. These changes can be seen in this [https://youtu.be/k4fjSZV_-OQ video] of a G-protein alpha subunit activation derived from common rats. | ||
=== Clinical Relevance === | === Clinical Relevance === | ||
| + | |||
== 3D Structures == | == 3D Structures == | ||
Revision as of 01:35, 29 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
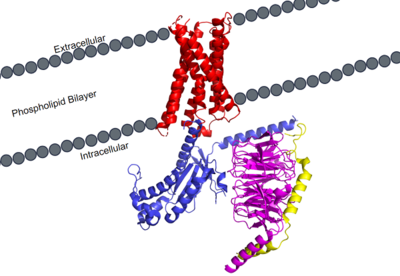
MRGPRX2 Human Itch G-Protein Coupled Receptor (GPCR)
| |||||||||||
References
- ↑ Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842. doi: 10.1038/nrd.2017.178. Epub, 2017 Oct 27. PMID:29075003 doi:http://dx.doi.org/10.1038/nrd.2017.178
- ↑ Basith S, Cui M, Macalino SJY, Park J, Clavio NAB, Kang S, Choi S. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front Pharmacol. 2018 Mar 9;9:128. doi: 10.3389/fphar.2018.00128. eCollection, 2018. PMID:29593527 doi:http://dx.doi.org/10.3389/fphar.2018.00128
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Cao C, Kang HJ, Singh I, Chen H, Zhang C, Ye W, Hayes BW, Liu J, Gumpper RH, Bender BJ, Slocum ST, Krumm BE, Lansu K, McCorvy JD, Kroeze WK, English JG, DiBerto JF, Olsen RHJ, Huang XP, Zhang S, Liu Y, Kim K, Karpiak J, Jan LY, Abraham SN, Jin J, Shoichet BK, Fay JF, Roth BL. Structure, function and pharmacology of human itch GPCRs. Nature. 2021 Dec;600(7887):170-175. doi: 10.1038/s41586-021-04126-6. Epub 2021, Nov 17. PMID:34789874 doi:http://dx.doi.org/10.1038/s41586-021-04126-6
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP. Structure, function and pharmacology of human itch receptor complexes. Nature. 2021 Dec;600(7887):164-169. doi: 10.1038/s41586-021-04077-y. Epub 2021, Nov 17. PMID:34789875 doi:http://dx.doi.org/10.1038/s41586-021-04077-y
- ↑ Kamato D, Thach L, Bernard R, Chan V, Zheng W, Kaur H, Brimble M, Osman N, Little PJ. Structure, Function, Pharmacology, and Therapeutic Potential of the G Protein, Galpha/q,11. Front Cardiovasc Med. 2015 Mar 24;2:14. doi: 10.3389/fcvm.2015.00014. eCollection, 2015. PMID:26664886 doi:http://dx.doi.org/10.3389/fcvm.2015.00014
- ↑ Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs--theoretical and experimental studies. Curr Med Chem. 2012;19(8):1090-109. doi: 10.2174/092986712799320556. PMID:22300046 doi:http://dx.doi.org/10.2174/092986712799320556
- ↑ Olivella M, Caltabiano G, Cordomi A. The role of Cysteine 6.47 in class A GPCRs. BMC Struct Biol. 2013 Mar 15;13:3. doi: 10.1186/1472-6807-13-3. PMID:23497259 doi:http://dx.doi.org/10.1186/1472-6807-13-3