Sandbox Reserved 1700
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
MRGPRX2 is a certain type of [https://proteopedia.org/wiki/index.php/G_protein-coupled_receptor GPCR] that is located in the cellular membranes of mast cells. | MRGPRX2 is a certain type of [https://proteopedia.org/wiki/index.php/G_protein-coupled_receptor GPCR] that is located in the cellular membranes of mast cells. | ||
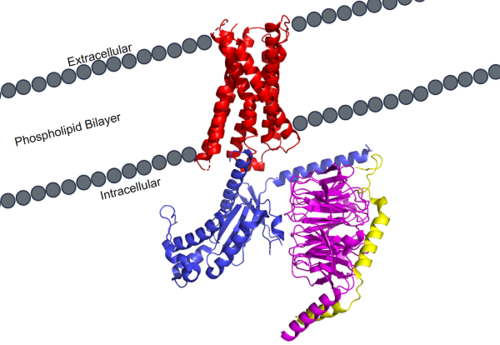
| - | [[Image:MRGPRX2_within_membrane.png|500px|right|thumb|Figure 1. MRGPRX2 as it sits within the cellular membrane. Phospholipid bilayer is represented by grey dots, with labeled cellular locations]] | + | [[Image:MRGPRX2_within_membrane.png|500px|right|thumb|'''Figure 1.''' MRGPRX2 as it sits within the cellular membrane. Phospholipid bilayer is represented by grey dots, with labeled cellular locations]] |
== Background == | == Background == | ||
| Line 20: | Line 20: | ||
=== G-Protein === | === G-Protein === | ||
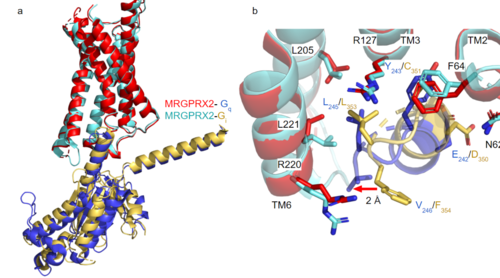
| - | [https://proteopedia.org/wiki/index.php/G_protein GTP-binding proteins], also known as G-proteins, are heterotrimeric complexes consisting of alpha, beta, and gamma subunits that interact with membrane receptor proteins. G-proteins are responsible transmitting extracellular signals into the cell upon activation. This activation happens when the alpha subunit of the G-protein binds GTP instead of GDP, and then disassociates from the rest of the protein, initiating the intracellular signaling cascade. There are different families of G-alpha subunits, Gαi, Gαs, Gα12/13, and Gαq <ref name="Kamato">PMID: 26664886</ref>. MRGPRX2 has been found to bind both Gαi and Gαq subunits with relatively no major structural changes between the two despite slightly different amino acids present <ref name= "Cao" /> <ref name= "Yang" />. Throughout this page, MGPRX2 is always shown with Gq. Figure 2a shows the overlay of MGPRX2 with either the Gq or Gi alpha subunit. Figure 2b shows the specific residues involved in the interface between the membrane receptor and G protein. The major difference between the two comes from one amino acid difference (valine on Gq versus phenylalanine on Gi) that pushes the Gi subunit 2Å away from the arginine residue on helix 6 of the transmembrane protein. This is the only major structural difference between Gq and Gi subunits. | + | [https://proteopedia.org/wiki/index.php/G_protein GTP-binding proteins], also known as G-proteins, are heterotrimeric complexes consisting of alpha, beta, and gamma subunits that interact with membrane receptor proteins. G-proteins are responsible transmitting extracellular signals into the cell upon activation. This activation happens when the alpha subunit of the G-protein binds GTP instead of GDP, and then disassociates from the rest of the protein, initiating the intracellular signaling cascade. There are different families of G-alpha subunits, Gαi, Gαs, Gα12/13, and Gαq <ref name="Kamato">PMID: 26664886</ref>. MRGPRX2 has been found to bind both Gαi and Gαq subunits with relatively no major structural changes between the two despite slightly different amino acids present <ref name= "Cao" /> <ref name= "Yang" />. Throughout this page, MGPRX2 is always shown with Gq. '''Figure 2a''' shows the overlay of MGPRX2 with either the Gq or Gi alpha subunit. '''Figure 2b''' shows the specific residues involved in the interface between the membrane receptor and G protein. The major difference between the two comes from one amino acid difference (valine on Gq versus phenylalanine on Gi) that pushes the Gi subunit 2Å away from the arginine residue on helix 6 of the transmembrane protein. This is the only major structural difference between Gq and Gi subunits. |
| - | [[Image:Gq and gi overlay.png|500px|center|thumb|Figure 2a. Overlay of MGPRX2-Gq (red-dark blue) and MGPRX2-Gi (cyan-yellow). Figure 2b. Important residues involved in the interface between MGPRX2 and Gq/ Gi subunits. Arrow pointing to the major difference between the interfaces, which comes from the final C-terminus residue on the G-alpha subunit. In Gq, there is a valine while in Gi, there is a phenylalanine. This pushes the Gi subunit 2Å away from the arginine residue on helix 6 of the transmembrane protein.]] | + | [[Image:Gq and gi overlay.png|500px|center|thumb|'''Figure 2a.''' Overlay of MGPRX2-Gq (red-dark blue) and MGPRX2-Gi (cyan-yellow). '''Figure 2b.''' Important residues involved in the interface between MGPRX2 and Gq/ Gi subunits. Arrow pointing to the major difference between the interfaces, which comes from the final C-terminus residue on the G-alpha subunit. In Gq, there is a valine while in Gi, there is a phenylalanine. This pushes the Gi subunit 2Å away from the arginine residue on helix 6 of the transmembrane protein.]] |
=== Novel Characteristics === | === Novel Characteristics === | ||
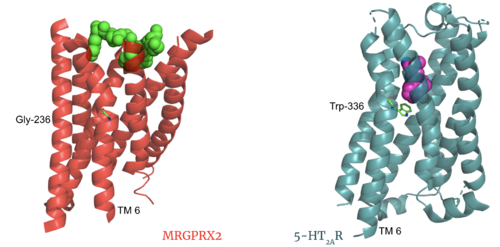
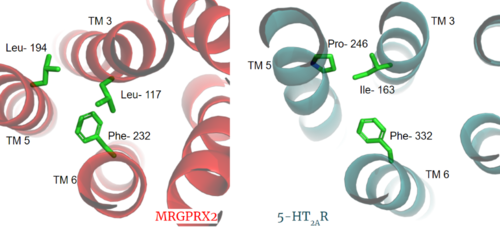
| - | The most important findings about the MRGPRX2 receptor are the differences between it and all other previously discovered GPCR's found in the A family. Many conserved structural motifs, characteristic of the A family receptors, are absent on MRGPRX2. These structural motif differences contribute to a membrane surface ligand binding rather than a ligand binding deep within the helices. To demonstrate this difference in depth binding, MRGPRX2 is compared to [https://proteopedia.org/wiki/index.php/Serotonin_receptor 5-HT2AR], another class A GPCR with more conserved structural motifs. | + | The most important findings about the MRGPRX2 receptor are the differences between it and all other previously discovered GPCR's found in the A family. Many conserved structural motifs, characteristic of the A family receptors, are absent on MRGPRX2. These structural motif differences contribute to a membrane surface ligand binding rather than a ligand binding deep within the helices ('''Figure 3'''). To demonstrate this difference in depth binding, MRGPRX2 is compared to [https://proteopedia.org/wiki/index.php/Serotonin_receptor 5-HT2AR], another class A GPCR with more conserved structural motifs. |
---- | ---- | ||
---- | ---- | ||
| - | [[Image:Screen Shot 2022-03-27 at 5.06.17 PM.png|500px|center|thumb|Figure 3. Comparison of ligand Cortistatin-14 binding in MRGPRX2 (left) and binding in 5HT2AR (right)]] | + | [[Image:Screen Shot 2022-03-27 at 5.06.17 PM.png|500px|center|thumb|'''Figure 3.''' Comparison of ligand Cortistatin-14 binding in MRGPRX2 (left) and binding in 5HT2AR (right)]] |
| Line 46: | Line 46: | ||
==== Toggle Switch ==== | ==== Toggle Switch ==== | ||
| - | Certain residues that lie in the ligand binding pocket can act as molecular switches to turn the GPCR “on” or “off” and are fittingly called toggle switches. Toggle switches in receptors are essential in interacting with the ligand upon binding, as they can then initiate the transmission of the molecular signal through the protein. Trp-336 has been known to act as this "iconic" toggle switch in GPCR’s of the A family <ref name="Trzaskowski">PMID: 22300046</ref>, and is also an important residue in another motif, known as the '''CWxP motif'''. However in MRGPRX2, the residue has been replaced to a <scene name='90/904305/Glycine_toggle_switch/7'>glycine</scene> <ref name="Cao">PMID: 34789874</ref> <ref name="Yang">PMID: 34789875</ref>. This is a significant modification to the receptor structure. By replacing the large tryptophan residue with a small glycine, the membrane helicies, especially helix 7 on which the toggle switch is found, can pack closer together. The ligands that interact with MRGPRX2 then bind much <scene name='90/904305/Glycine_toggle_switch_and_cor/1'>closer to the surface</scene> of the receptor, as opposed to deeper within the helices. This significant change can be visualized in | + | Certain residues that lie in the ligand binding pocket can act as molecular switches to turn the GPCR “on” or “off” and are fittingly called toggle switches. Toggle switches in receptors are essential in interacting with the ligand upon binding, as they can then initiate the transmission of the molecular signal through the protein. Trp-336 has been known to act as this "iconic" toggle switch in GPCR’s of the A family <ref name="Trzaskowski">PMID: 22300046</ref>, and is also an important residue in another motif, known as the '''CWxP motif'''. However in MRGPRX2, the residue has been replaced to a <scene name='90/904305/Glycine_toggle_switch/7'>glycine</scene> <ref name="Cao">PMID: 34789874</ref> <ref name="Yang">PMID: 34789875</ref>. This is a significant modification to the receptor structure. By replacing the large tryptophan residue with a small glycine, the membrane helicies, especially helix 7 on which the toggle switch is found, can pack closer together. The ligands that interact with MRGPRX2 then bind much <scene name='90/904305/Glycine_toggle_switch_and_cor/1'>closer to the surface</scene> of the receptor, as opposed to deeper within the helices. This significant change can be visualized in '''Figure 3'''. This significantly changes what structures of ligands are able to interact with this receptor and therefore what types of molecules can activate the Human Itch GPCR. More details about what kinds of ligands bind to this receptor are discussed later. |
==== Sodium Site ==== | ==== Sodium Site ==== | ||
| Line 65: | Line 65: | ||
==== DRY/ ERC Motif ==== | ==== DRY/ ERC Motif ==== | ||
MRGPRX2 has an <scene name='90/904306/Erc_motif_3/1'>ERC Motif</scene> rather than the typically [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY Motif]. The amino acid residue shift from TYR-174 to CYS-128 has spatial arrangement implications where the helices are more compact in MRGPRX2 without the TYR to physically push the TMP helices apart. | MRGPRX2 has an <scene name='90/904306/Erc_motif_3/1'>ERC Motif</scene> rather than the typically [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY Motif]. The amino acid residue shift from TYR-174 to CYS-128 has spatial arrangement implications where the helices are more compact in MRGPRX2 without the TYR to physically push the TMP helices apart. | ||
| - | [[Image:Screen Shot 2022-03-15 at 10.23.20 AM.png|200px|left|thumb|ERC Motif]] | + | [[Image:Screen Shot 2022-03-15 at 10.23.20 AM.png|200px|left|thumb|'''Figure 4.''' ERC Motif]] |
| Line 91: | Line 91: | ||
==== Disulfide Bonds ==== | ==== Disulfide Bonds ==== | ||
The MRGPRX2 disulfide bond is between <scene name='90/904305/Disulfide_bond/1'>Cys-168 and Cys-180</scene> on TM helices 5 and 4, respectively. In other class A GPCRs, this disulfide bond is between and extracellular loop (ECL2) and a TM helix (TM3). For example, the <scene name='90/904306/5ht2a_disulfide/2'>serotonin GPCR</scene> shows this disulfide bond between the ECL2 and TM3. This different disulfide bond location contributes to surface level binding of ligands. | The MRGPRX2 disulfide bond is between <scene name='90/904305/Disulfide_bond/1'>Cys-168 and Cys-180</scene> on TM helices 5 and 4, respectively. In other class A GPCRs, this disulfide bond is between and extracellular loop (ECL2) and a TM helix (TM3). For example, the <scene name='90/904306/5ht2a_disulfide/2'>serotonin GPCR</scene> shows this disulfide bond between the ECL2 and TM3. This different disulfide bond location contributes to surface level binding of ligands. | ||
| - | [[Image:Screen Shot 2022-03-27 at 5.45.52 PM.png|300px|center|thumb|Overlay of the 5HT2AR and MRGPRX2 TMP for comparison of disulfide bond location.]] | + | [[Image:Screen Shot 2022-03-27 at 5.45.52 PM.png|300px|center|thumb|'''Figure 5.''' Overlay of the 5HT2AR and MRGPRX2 TMP for comparison of disulfide bond location.]] |
| Line 106: | Line 106: | ||
=== Ligands === | === Ligands === | ||
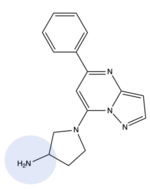
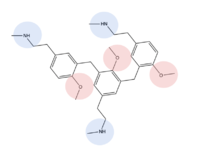
MRGPRX2 binds a wide range of small molecule and peptide ligands. These ligands have positively charged regions which can interact with the negative binding pocket, sub-pocket 1. Some of these larger ligands also have a hydrophobic region which can interact with sub-pocket 2. MRGPRX2 has been known to interact with [https://pubchem.ncbi.nlm.nih.gov/compound/PAMP-12-_human_-porcine#section=Structures PAMP-12], [https://pubchem.ncbi.nlm.nih.gov/compound/44208884 Cortistatin-14], [https://pubchem.ncbi.nlm.nih.gov/compound/118797323 C48/80], and [https://pubchem.ncbi.nlm.nih.gov/compound/95882507 Zinc-3573]<ref name="Yang">PMID: 34789875</ref>,<ref name="Cao">PMID: 34789874</ref>. | MRGPRX2 binds a wide range of small molecule and peptide ligands. These ligands have positively charged regions which can interact with the negative binding pocket, sub-pocket 1. Some of these larger ligands also have a hydrophobic region which can interact with sub-pocket 2. MRGPRX2 has been known to interact with [https://pubchem.ncbi.nlm.nih.gov/compound/PAMP-12-_human_-porcine#section=Structures PAMP-12], [https://pubchem.ncbi.nlm.nih.gov/compound/44208884 Cortistatin-14], [https://pubchem.ncbi.nlm.nih.gov/compound/118797323 C48/80], and [https://pubchem.ncbi.nlm.nih.gov/compound/95882507 Zinc-3573]<ref name="Yang">PMID: 34789875</ref>,<ref name="Cao">PMID: 34789874</ref>. | ||
| - | [[Image:Image-Screen Shot 2022-03-28 at 8.21.55 PM.png|150px|left|thumb|Zinc-3573]][[Image:80.png|200px|center|thumb|C48/80]] | + | [[Image:Image-Screen Shot 2022-03-28 at 8.21.55 PM.png|150px|left|thumb|'''Figure 6.''' Zinc-3573]][[Image:80.png|200px|center|thumb|'''Figure 7.''' C48/80]] |
| - | [[Image:Screen Shot 2022-03-28 at 8.24.59 PM.png|300px|left|thumb|PAMP-12]][[Image:Screen Shot 2022-03-28 at 8.25.58 PM.png|300px|center|thumb|Cortistatin-14]] | + | [[Image:Screen Shot 2022-03-28 at 8.24.59 PM.png|300px|left|thumb|'''Figure 8.''' PAMP-12]][[Image:Screen Shot 2022-03-28 at 8.25.58 PM.png|300px|center|thumb|'''Figure 9.''' Cortistatin-14]] |
| Line 121: | Line 121: | ||
=== Before Activation === | === Before Activation === | ||
The culmination of different motifs observed in MRGPRX2 compared to other class A GPCRs leads to external membrane ligand binding. The MRGPRX2 GPCR undergoes a much smaller conformational change upon ligand binding compared to other Class A GPCRs due to surface level binding versus deep helix binding. This contrast is represented in the photo below to show the unbound and bound transmembrane proteins of MRGPRX2 and 5-HT2A. | The culmination of different motifs observed in MRGPRX2 compared to other class A GPCRs leads to external membrane ligand binding. The MRGPRX2 GPCR undergoes a much smaller conformational change upon ligand binding compared to other Class A GPCRs due to surface level binding versus deep helix binding. This contrast is represented in the photo below to show the unbound and bound transmembrane proteins of MRGPRX2 and 5-HT2A. | ||
| - | [[Image:Screen Shot 2022-03-282 at 6.59.44 PM.png|500px|center|thumb|Overlay of unbound (transparent) and bound (opaque) transmembrane proteins of both MRGRPX2 (left) and 5-HT2AR (right).]] | + | [[Image:Screen Shot 2022-03-282 at 6.59.44 PM.png|500px|center|thumb|'''Figure 10.''' Overlay of unbound (transparent) and bound (opaque) transmembrane proteins of both MRGRPX2 (left) and 5-HT2AR (right).]] |
=== After Activation === | === After Activation === | ||
Revision as of 13:06, 29 March 2022
MRGPRX2 Human Itch G-Protein Coupled Receptor (GPCR)
| |||||||||||
References
- ↑ Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842. doi: 10.1038/nrd.2017.178. Epub, 2017 Oct 27. PMID:29075003 doi:http://dx.doi.org/10.1038/nrd.2017.178
- ↑ Basith S, Cui M, Macalino SJY, Park J, Clavio NAB, Kang S, Choi S. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front Pharmacol. 2018 Mar 9;9:128. doi: 10.3389/fphar.2018.00128. eCollection, 2018. PMID:29593527 doi:http://dx.doi.org/10.3389/fphar.2018.00128
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Cao C, Kang HJ, Singh I, Chen H, Zhang C, Ye W, Hayes BW, Liu J, Gumpper RH, Bender BJ, Slocum ST, Krumm BE, Lansu K, McCorvy JD, Kroeze WK, English JG, DiBerto JF, Olsen RHJ, Huang XP, Zhang S, Liu Y, Kim K, Karpiak J, Jan LY, Abraham SN, Jin J, Shoichet BK, Fay JF, Roth BL. Structure, function and pharmacology of human itch GPCRs. Nature. 2021 Dec;600(7887):170-175. doi: 10.1038/s41586-021-04126-6. Epub 2021, Nov 17. PMID:34789874 doi:http://dx.doi.org/10.1038/s41586-021-04126-6
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP. Structure, function and pharmacology of human itch receptor complexes. Nature. 2021 Dec;600(7887):164-169. doi: 10.1038/s41586-021-04077-y. Epub 2021, Nov 17. PMID:34789875 doi:http://dx.doi.org/10.1038/s41586-021-04077-y
- ↑ Kamato D, Thach L, Bernard R, Chan V, Zheng W, Kaur H, Brimble M, Osman N, Little PJ. Structure, Function, Pharmacology, and Therapeutic Potential of the G Protein, Galpha/q,11. Front Cardiovasc Med. 2015 Mar 24;2:14. doi: 10.3389/fcvm.2015.00014. eCollection, 2015. PMID:26664886 doi:http://dx.doi.org/10.3389/fcvm.2015.00014
- ↑ Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs--theoretical and experimental studies. Curr Med Chem. 2012;19(8):1090-109. doi: 10.2174/092986712799320556. PMID:22300046 doi:http://dx.doi.org/10.2174/092986712799320556
- ↑ Olivella M, Caltabiano G, Cordomi A. The role of Cysteine 6.47 in class A GPCRs. BMC Struct Biol. 2013 Mar 15;13:3. doi: 10.1186/1472-6807-13-3. PMID:23497259 doi:http://dx.doi.org/10.1186/1472-6807-13-3