This is a default text for your page GeorgeAnna/VKOR. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
Biological Role
(VKOR) is an enzyme that reduces (KO) to Vitamin K and ultimately Vitamin K hydroquinone (KH2) (cite fig 1). VKOR is a 4-helix transmembrane protein spanning the endoplasmic reticulum . One of its primary roles is to assist in blood coagulation through KH2 regeneration. KH2 is a necessary cofactor for the γ-carboxylase enzyme that activates several coagulation factors.
Structural characterization of VKOR has been difficult due to its in vitro instability. Recently, a series of atomic structures have been determined utilizing anticoagulant stabilization and VKOR-like homologs.
Author's Notes
Crystal structures of VKOR were captured with a bound substrate (KO) or vitamin K antagonist (VKA) (PDB Figure)  . VKA substrates utilized were anticoagulants, namely Warfarin, Brodifacoum, Phenindione, and Chlorophacinone. Second, VKOR-like homologs were utilized to aid in structure classification. Homologs refer to specific cysteine residues that have been mutated to serine to facilitate capturing a stable conformation state. Homologs were mainly isolated from human VKOR with some isolated from the pufferfish Takifugu rubripes. Furthermore, all of the structures used have been processed to remove a beta barrel at the south end of VKOR that served no purpose in function of the enzyme. This also allowed for the residue numbering to be reassigned and more closely replicate the human VKOR.
. VKA substrates utilized were anticoagulants, namely Warfarin, Brodifacoum, Phenindione, and Chlorophacinone. Second, VKOR-like homologs were utilized to aid in structure classification. Homologs refer to specific cysteine residues that have been mutated to serine to facilitate capturing a stable conformation state. Homologs were mainly isolated from human VKOR with some isolated from the pufferfish Takifugu rubripes. Furthermore, all of the structures used have been processed to remove a beta barrel at the south end of VKOR that served no purpose in function of the enzyme. This also allowed for the residue numbering to be reassigned and more closely replicate the human VKOR.
Structural Highlights
Active Site
Within the four transmembrane helices lies the . The active site is comprised of a hydrophobic pocket containing two hydrophilic residues, A80 and Y139, that interact with substrates and ligands alike. The hydrophobic pocket provides specificity to the region while the hydrophilic residues have potential to hydrogen bond, allowing recognition and increasing specificity as well. Slightly above the active site is a crucial disulfide bridge that provides stabilization when a substrate is bound. This bridge occurs between C132 and C135, recurrent residues that continually aid in VKOR function.
The active site plays a vital role in binding of any substrate or ligand to the VKOR. Upon binding, the VKOR will transition into a that will allow its catalytic mechanism to commence.
Cap Domain
Anchor
Function: Method of Coagulation
Brief Overview
In its resting state, VKOR is in an , prepared for a substrate to bind. Once the substrate binds, this will induce the closed conformation of VKOR, where the catalytic mechanism will activate Vitamin K via reactive cysteine residues. Vitamin K will then be released from the binding pocket for use in the body, and VKOR will return to the open conformation once again. The enzyme will then reset into its reactive state to prep for another molecule of Vitamin K to bind.
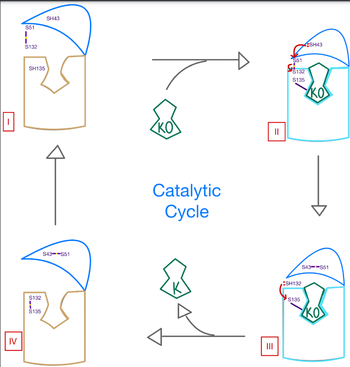
Catalytic Mechanism
The catalytic mechanism of VKOR is a critical part of its overall function in the body. Highly regulated enzymatic activity through the reactivity of catalytic cysteines allows VKOR to properly activate Vitamin K for its use in the body. The enzyme begins in , where it's in the open conformation with the cap domain open to allow in a substrate to bind to the active site. Once a substrate binds, the cap domain is initiated into the closed conformation. VKOR is now in . To further stabilize the closed conformation with the substrate bound, the cap domain helps initiate a catalytic reaction of cysteines to break the disulfide bridge that was stabilizing stage 1. Free cysteines are now available that provide strong stabilization of the closed conformation through interactions with the cap domain and the bound substrate. This puts the enzyme in , where the catalytic free cysteines react to form a new disulfide bridge, releasing the activated substrate into the blood stream to promote anticoagulation. With two stable disulfide bridges and VKOR unbound, the enzyme is now in its final, unreactive . VKOR must undergo conformational changes to return to Stage 1 and restart the catalytic process to activate Vitamin K again.
Disease and Treatment
Afflictions
Inhibition
The most inexpensive and common way to treat blood clotting is through the VKOR inhibitor, . Warfarin is able to do so by outcompeting KO. It will enter the binding pocket of VKOR, creating strong hydrogen bonds with the active site.
Mutations
Some key that can be detrimental to the VKOR structure are mutations of the . The two main residues, N80 and Y139, can be mutated to A80 and F139 creating a decrease in recognition and stabilization.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.