We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1726
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
=== Domains === | === Domains === | ||
==== Three Helix Bundle-like Domain ==== | ==== Three Helix Bundle-like Domain ==== | ||
| - | The <scene name='90/904331/Thb_like_domain/ | + | The <scene name='90/904331/Thb_like_domain/2'>Three Helix Bundle-like Domain</scene> mainly has a structural function overall as it interacts with the tumor necrosis factor-like domain upon ligand binding. The three helix bundle-like domain's α-helix interacts with the helix α-1' and β strand A-1' on the Tumor Necrosis Factor-like domain. This outermost region of the extracellular ligand-binding domain undergoes rigorous structural reorientation upon ligand binding. THB is primarily involved in the dimerization motif of ALK, which dimerizes upon ligand binding. <ref name="Reshetnyak" /> |
==== Poly-Glycine Domain ==== | ==== Poly-Glycine Domain ==== | ||
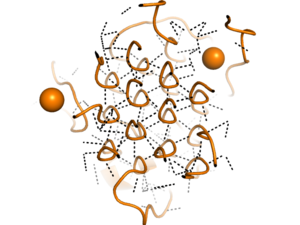
| - | [[Image:glycinehelicesorange.png|300 px|right|thumb|Figure 2. Rare Glycine helices on Anaplastic Lymphoma Kinase]]Located between the three helix bundle-like domain and the tumor necrosis factor-like domain, the <scene name='90/904331/Polyg_region1/ | + | [[Image:glycinehelicesorange.png|300 px|right|thumb|Figure 2. Rare Glycine helices on Anaplastic Lymphoma Kinase]]Located between the three helix bundle-like domain and the tumor necrosis factor-like domain, the <scene name='90/904331/Polyg_region1/3'>Poly-Glycine Region</scene> has an important structural role. The poly-Glycine domain also has a rare and unique structure of left-handed glycine helices with hexagonal hydrogen bonding shown in Figure 2. These 14 glycine helices are unique to ALK's function among other tyrosine kinases, as these types of structures on the binding domain are not present. These helices are rigid structures, providing a strong anchor for the ligand binding site while the other domains undergo drastic conformational rearrangements.<ref name="Reshetnyak" /> |
==== Tumor-Necrosis Factor-like Domain ==== | ==== Tumor-Necrosis Factor-like Domain ==== | ||
| - | The <scene name='90/904331/Tnf_like_domain/ | + | The <scene name='90/904331/Tnf_like_domain/2'>Tumor Necrosis Factor-like Domain</scene> interacts with the three helix bundle-like domain to begin the conformational changes associated with ligand binding. It is located in approximately the midregion of the extracellular region, bridging the gap between the poly-glycine domain and the epidermal growth factor-like domain. This domain also assists in mediating ligand binding with the epidermal growth factor-like domain. In ligand-binding, as previously stated, this domain interacts heavily with the THB to undergo critical conformation changes necessary for dimerization and ligand recognition. <ref name="Reshetnyak" /> |
==== Epidermal Growth Factor-like Domain ==== | ==== Epidermal Growth Factor-like Domain ==== | ||
| - | The <scene name='90/904331/Egf_like_domain/ | + | The <scene name='90/904331/Egf_like_domain/2'>Epidermal Growth Factor-like Domain</scene> is very malleable and repositioning of this domain is essential for activation of the protein. This domain is able to undergo conformational changes with the ligand bound and when in contact with the tumor necrosis factor-like domain. The interface between the EGF-like and TNF-like domains are primarily hydrophobic residues, which enable their flexibility with regards to one another. The main motifs that are apart of the EGF-like domain are major and minor β-hairpins, which are stabilized by 3 conserved disulfide bridges. <ref name="Reshetnyak" /> |
=== Binding Site === | === Binding Site === | ||
This site doesn't start out surrounding the [https://en.wikipedia.org/wiki/Ligand_(biochemistry) ligand], instead the proximity of the ligand allows [https://en.wikipedia.org/wiki/Conformational_change conformational changes] across the protein. The ligands for ALK both have highly positively charged faces that interact with the TNF-like region, the primary ligand-binding site on the extracellular region<ref name="Li" />. [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) Salt bridges] between the positively charged residues on the ligand and negatively charged residues on the receptor form are formed as the ligand approaches connecting the ligand with the receptor. Three of these salt bridges occur between <scene name='90/904331/Salt_bridge_859_140/2'>E859 and R140</scene>, <scene name='90/904331/Salt_bridge_974_136/3'>E974 and R136</scene>, and <scene name='90/904331/Salt_bridge_978_123_133/2'>E978 with both R123 and R133</scene>. These strong ionic interactions allow the drastic conformational changes in the extracellular domain that induce the signaling pathway. <ref name="Reshetnyak" /> | This site doesn't start out surrounding the [https://en.wikipedia.org/wiki/Ligand_(biochemistry) ligand], instead the proximity of the ligand allows [https://en.wikipedia.org/wiki/Conformational_change conformational changes] across the protein. The ligands for ALK both have highly positively charged faces that interact with the TNF-like region, the primary ligand-binding site on the extracellular region<ref name="Li" />. [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) Salt bridges] between the positively charged residues on the ligand and negatively charged residues on the receptor form are formed as the ligand approaches connecting the ligand with the receptor. Three of these salt bridges occur between <scene name='90/904331/Salt_bridge_859_140/2'>E859 and R140</scene>, <scene name='90/904331/Salt_bridge_974_136/3'>E974 and R136</scene>, and <scene name='90/904331/Salt_bridge_978_123_133/2'>E978 with both R123 and R133</scene>. These strong ionic interactions allow the drastic conformational changes in the extracellular domain that induce the signaling pathway. <ref name="Reshetnyak" /> | ||
Revision as of 22:17, 12 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
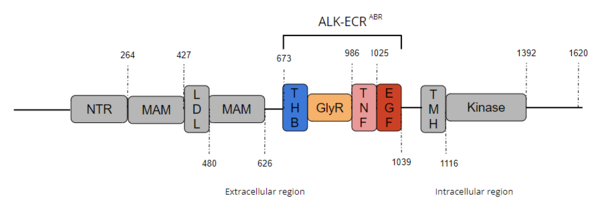
Anaplastic Lymphoma Kinase Extracellular Region
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018 Nov 2;19(11). pii: ijms19113448. doi: 10.3390/ijms19113448. PMID:30400214 doi:http://dx.doi.org/10.3390/ijms19113448
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Reshetnyak AV, Rossi P, Myasnikov AG, Sowaileh M, Mohanty J, Nourse A, Miller DJ, Lax I, Schlessinger J, Kalodimos CG. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021 Dec;600(7887):153-157. doi: 10.1038/s41586-021-04140-8. Epub 2021, Nov 24. PMID:34819673 doi:http://dx.doi.org/10.1038/s41586-021-04140-8
- ↑ 3.0 3.1 Li T, Stayrook SE, Tsutsui Y, Zhang J, Wang Y, Li H, Proffitt A, Krimmer SG, Ahmed M, Belliveau O, Walker IX, Mudumbi KC, Suzuki Y, Lax I, Alvarado D, Lemmon MA, Schlessinger J, Klein DE. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature. 2021 Dec;600(7887):148-152. doi: 10.1038/s41586-021-04141-7. Epub 2021, Nov 24. PMID:34819665 doi:http://dx.doi.org/10.1038/s41586-021-04141-7
- ↑ 4.0 4.1 4.2 4.3 Borenas M, Umapathy G, Lai WY, Lind DE, Witek B, Guan J, Mendoza-Garcia P, Masudi T, Claeys A, Chuang TP, El Wakil A, Arefin B, Fransson S, Koster J, Johansson M, Gaarder J, Van den Eynden J, Hallberg B, Palmer RH. ALK ligand ALKAL2 potentiates MYCN-driven neuroblastoma in the absence of ALK mutation. EMBO J. 2021 Feb 1;40(3):e105784. doi: 10.15252/embj.2020105784. Epub 2021 Jan 7. PMID:33411331 doi:http://dx.doi.org/10.15252/embj.2020105784
- ↑ 5.0 5.1 Della Corte CM, Viscardi G, Di Liello R, Fasano M, Martinelli E, Troiani T, Ciardiello F, Morgillo F. Role and targeting of anaplastic lymphoma kinase in cancer. Mol Cancer. 2018 Feb 19;17(1):30. doi: 10.1186/s12943-018-0776-2. PMID:29455642 doi:http://dx.doi.org/10.1186/s12943-018-0776-2