We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1710
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Function == | == Function == | ||
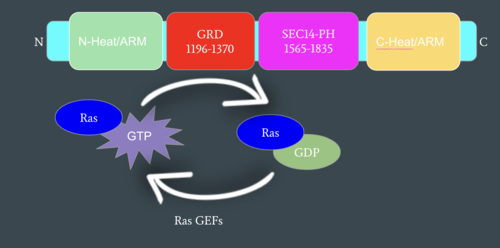
[[Image:mechanismofRas.png|500 px|left|thumb|Figure 2: Mechanism of Ras Regulation by Neurofibromin. One chain of the homodimer is represented in cyan with its important domains highlighted. Ras Guanine Nucleotide Exchange Factors (RasGEFs) are shown catalyzing the transition from an active to an inactive Ras molecule and Neurofibromin is shown catalyzing the reverse reaction]] | [[Image:mechanismofRas.png|500 px|left|thumb|Figure 2: Mechanism of Ras Regulation by Neurofibromin. One chain of the homodimer is represented in cyan with its important domains highlighted. Ras Guanine Nucleotide Exchange Factors (RasGEFs) are shown catalyzing the transition from an active to an inactive Ras molecule and Neurofibromin is shown catalyzing the reverse reaction]] | ||
| - | Neurofibromin functions as a tumor suppressor protein.<ref name="Trovó-Marqui">PMID:16813595</ref> It prevents cell growth by turning off [https://en.wikipedia.org/wiki/Ras_GTPase Ras] which in its active state, stimulates cell growth and division (Figure 2). Ras is a small, monomeric [https://en.wikipedia.org/wiki/GTPase GTPase] that binds to [https://en.wikipedia.org/wiki/Guanosine_triphosphate GTP] and hydrolyzes it to [https://en.wikipedia.org/wiki/Guanosine_diphosphate GDP]. When Ras is active, it subsequently activates other proteins that stimulate cell growth and proliferation. Ras is membrane-bound and interacts with Neurofibromin, a cytoplasmic protein, in the open conformation of Neurofibromin. Neurofibromin is brought to the membrane to associate with Ras by [https://en.wikipedia.org/wiki/SPRED1 SPRED1]. [https://medlineplus.gov/genetics/gene/spred1/ SPRED1] is a protein that helps regulate the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway Ras/MAPK signaling pathway] responsible for the growth and proliferation of cells. Binding of SPRED1 to [https://en.wikipedia.org/wiki/RAF_kinase Raf] in the MAPK signaling pathway blocks the activation of Raf, halts the rest of the pathway, and stops cell growth and proliferation. Unlike Ras, Neurofibromin can interact with SPRED1 in both the open and closed conformations.<ref name="Naschberger">PMID:34707296</ref> The interaction between Neurofibromin and Ras is activated via an [https://en.wikipedia.org/wiki/Arginine_finger Arginine finger] (Arg1276) present in the GRD domain of Neurofibromin. Arg1276 is only accessible for binding when the GRD and Sec14-PH domains are rotated into the open conformation. Ras binds to the arginine finger of Neurofibromin with its switch regions 1 and 2.<ref name="Trovó-Marqui">PMID:16813595</ref> When Arg1276 is able to associate with Ras, Neurofibromin downregulates the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway Ras signaling pathway] by speeding up Ras's GTPase activity, hydrolyzing the GTP associated with Ras to GDP. In its GDP bound state, Ras is inactive and cell growth and division is inhibited. <ref name="Naschberger">PMID:34707296</ref> The positively charged arginine finger stabilizes the transition state for GTP hydrolysis by neutralizing the negative charges on GTP, which helps increase the speed of hydrolysis, fulfilling its catalytic function. <ref name="Trovó-Marqui">PMID:16813595</ref> | + | Neurofibromin functions as a tumor suppressor protein.<ref name="Trovó-Marqui">PMID:16813595</ref> It prevents cell growth by turning off [https://en.wikipedia.org/wiki/Ras_GTPase Ras] which in its active state, stimulates cell growth and division (Figure 2). Ras is a small, monomeric [https://en.wikipedia.org/wiki/GTPase GTPase] that binds to [https://en.wikipedia.org/wiki/Guanosine_triphosphate GTP] and hydrolyzes it to [https://en.wikipedia.org/wiki/Guanosine_diphosphate GDP]. When Ras is active, it subsequently activates other proteins that stimulate cell growth and proliferation. Ras is membrane-bound and interacts with Neurofibromin, a cytoplasmic protein, in the open conformation of Neurofibromin. Neurofibromin is brought to the membrane to associate with Ras by [https://en.wikipedia.org/wiki/SPRED1 SPRED1]. [https://medlineplus.gov/genetics/gene/spred1/ SPRED1] is a protein that helps regulate the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway Ras/MAPK signaling pathway] responsible for the growth and proliferation of cells. Binding of SPRED1 to [https://en.wikipedia.org/wiki/RAF_kinase Raf] in the MAPK signaling pathway blocks the activation of Raf, halts the rest of the pathway, and stops cell growth and proliferation. Unlike Ras, Neurofibromin can interact with SPRED1 in both the open and closed conformations.<ref name="Naschberger">PMID:34707296</ref> The interaction between Neurofibromin and Ras is activated via an [https://en.wikipedia.org/wiki/Arginine_finger Arginine finger] (Arg1276) present in the GRD domain of Neurofibromin. Arg1276 is only accessible for binding when the GRD and Sec14-PH domains are rotated into the open conformation. Ras binds to the arginine finger of Neurofibromin with its switch regions 1 and 2.<ref name="Trovó-Marqui">PMID:16813595</ref> When Arg1276 is able to associate with Ras, Neurofibromin downregulates the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway Ras signaling pathway] by speeding up Ras's GTPase activity, hydrolyzing the GTP associated with Ras to GDP. In its GDP bound state, Ras is inactive and cell growth and division is inhibited. <ref name="Naschberger">PMID:34707296</ref> The positively charged arginine finger stabilizes the transition state for GTP hydrolysis by neutralizing the negative charges on GTP, which helps increase the speed of hydrolysis, fulfilling its catalytic function. <ref name="Trovó-Marqui">PMID:16813595</ref> |
| - | + | ||
== Structure == | == Structure == | ||
[[Image:Neurofibromin_Cartoon_Domains.jpg|500 px|right|thumb|Figure 3: Neurofibromin Important Domains in one chain of the homodimer]] | [[Image:Neurofibromin_Cartoon_Domains.jpg|500 px|right|thumb|Figure 3: Neurofibromin Important Domains in one chain of the homodimer]] | ||
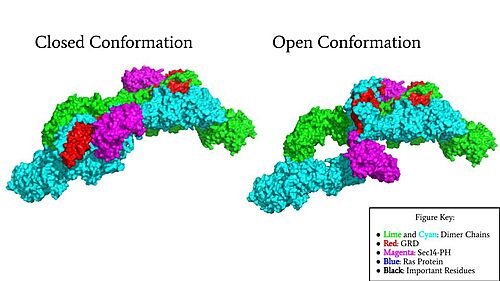
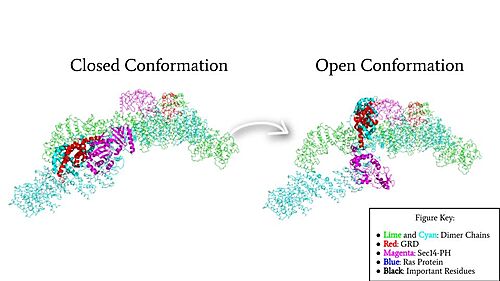
| - | Neurofibromin is a <scene name='90/904315/Homodimer/5'>homodimer</scene> made up of two identical chains. Neurofibromin has two conformations, open and closed. Shifting between these controls neurofibromin's ability to associate with Ras and perform its function of Ras regulation. The transformation between the overall closed and open conformations transitions it from an active to inactive state. There are <scene name='90/904315/Sec14ph_and_grd_domain/1'>two important domains</scene> involved in the transition between the open and closed conformations, the <scene name='90/904316/Grd_domains/2'>GRD</scene> domain and the <scene name='90/904315/Sec14ph_domain/3'>Sec14-PH</scene> domain. The GRD and the Sec14-PH domain are centrally linked | + | Neurofibromin is a <scene name='90/904315/Homodimer/5'>homodimer</scene> made up of two identical chains. Neurofibromin has two conformations, open and closed. Shifting between these controls neurofibromin's ability to associate with Ras and perform its function of Ras regulation. The transformation between the overall closed and open conformations transitions it from an active to inactive state. There are <scene name='90/904315/Sec14ph_and_grd_domain/1'>two important domains</scene> involved in the transition between the open and closed conformations, the <scene name='90/904316/Grd_domains/2'>GRD</scene> domain and the <scene name='90/904315/Sec14ph_domain/3'>Sec14-PH</scene> domain. The GRD and the Sec14-PH domain are centrally linked by an asymmetric, homodimeric core of four [https://en.wikipedia.org/wiki/Armadillo_repeat ARM] repeats and 27 [https://en.wikipedia.org/wiki/HEAT_repeat HEAT] repeats (Figure 3). The GRD and Sec14-PH domains extend out from the <scene name='90/904315/N-heat_arm/1'>N-HEAT/ARM</scene> core and then return to the <scene name='90/904315/C-heat_arm/1'>C-HEAT/ARM</scene> core. The orientation of the GRD and Sec-14 in relation to the N-HEAT/ARM and C-Heat/ARM determine what conformation neurofibromin is in. Although neurofibromin is a homodimer with two identical protomers, only one protomer has its GRD and Sec14-PH domains rotated into the <scene name='90/904315/Open_conformation/4'>open conformation</scene>.<ref name="Naschberger">PMID:34707296</ref> |
== Conformational States == | == Conformational States == | ||
Current revision
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Human Neurofibromin - The Tumor Suppressor Gene
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Trovo-Marqui AB, Tajara EH. Neurofibromin: a general outlook. Clin Genet. 2006 Jul;70(1):1-13. doi: 10.1111/j.1399-0004.2006.00639.x. PMID:16813595 doi:http://dx.doi.org/10.1111/j.1399-0004.2006.00639.x
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Naschberger A, Baradaran R, Rupp B, Carroni M. The structure of neurofibromin isoform 2 reveals different functional states. Nature. 2021 Nov;599(7884):315-319. doi: 10.1038/s41586-021-04024-x. Epub 2021, Oct 27. PMID:34707296 doi:http://dx.doi.org/10.1038/s41586-021-04024-x
- ↑ 3.0 3.1 Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ 4.0 4.1 4.2 Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015 May;15(5):290-301. doi: 10.1038/nrc3911. Epub 2015 Apr 16. PMID:25877329 doi:http://dx.doi.org/10.1038/nrc3911
- ↑ 5.0 5.1 5.2 Abramowicz A, Gos M. Neurofibromin in neurofibromatosis type 1 - mutations in NF1gene as a cause of disease. Dev Period Med. 2014 Jul-Sep;18(3):297-306. PMID:25182393
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365