BASIL2022GV3R8E
From Proteopedia
| Line 24: | Line 24: | ||
== Conclusions/Future Direction == | == Conclusions/Future Direction == | ||
Conclusions: | Conclusions: | ||
| + | |||
- D-Glucose exhibited a specific activity of 4.22 ug/mL | - D-Glucose exhibited a specific activity of 4.22 ug/mL | ||
| + | |||
- Specific activity: D-Glucose >> D-Galactose > D-Fructose > Lactose > D-Ribose | - Specific activity: D-Glucose >> D-Galactose > D-Fructose > Lactose > D-Ribose | ||
| + | |||
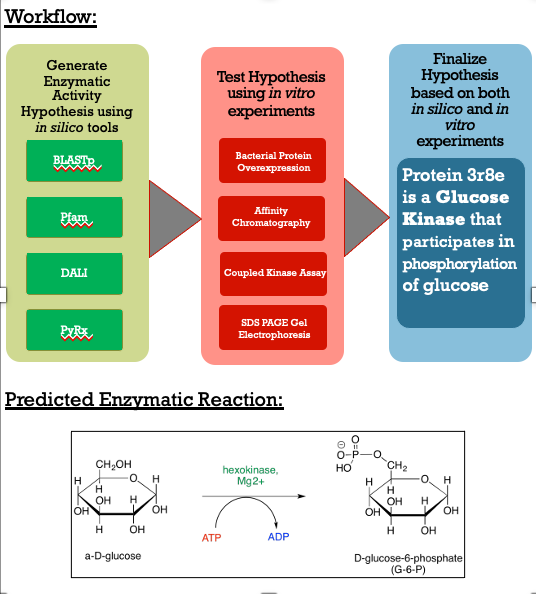
- Protein 3r8e is a Glucose Kinase | - Protein 3r8e is a Glucose Kinase | ||
| + | |||
- Glucose is phosphorylated by our protein | - Glucose is phosphorylated by our protein | ||
| + | |||
Future Direction: | Future Direction: | ||
| + | |||
- Verify experimental results | - Verify experimental results | ||
| + | |||
- Publish results for further application | - Publish results for further application | ||
| + | |||
- Authenticate protein function using further kinase characterization protocols | - Authenticate protein function using further kinase characterization protocols | ||
Revision as of 00:31, 21 April 2022
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
3. Blastp [Internet]. Bethesda (MD): Natiobal Library of Medicine (US), National Center for Biotechnology Information; 2004- [cited 2022 March]. Available from: (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)
4. BASIL. https://basilbiochem.github.io/basil/
5. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
6. Small- Molecule Library Screening by Docking with PyRx. .Dallakyan S, Olson AJ Methods Mol Biol. 2015;1263:243-50. The full-text is available at https://www.researchgate.net/publications/2739554875. Small-Molecule Library Screening by Docking with PyRx.
7. Pfam: The Protein families database in 2021 J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
8. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky