BASIL2022GV3R8E
From Proteopedia
| Line 13: | Line 13: | ||
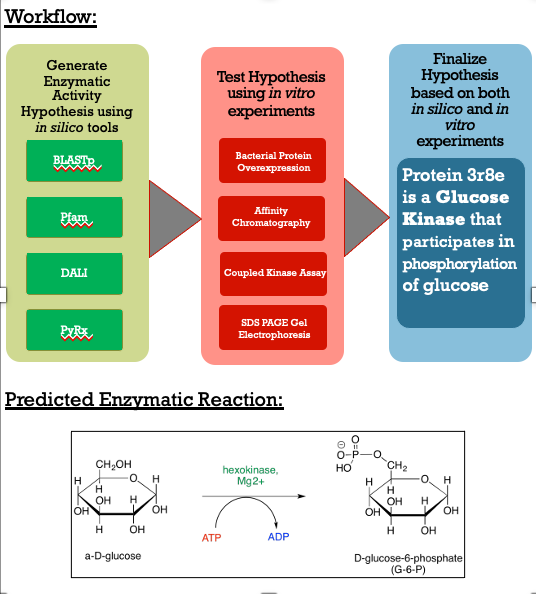
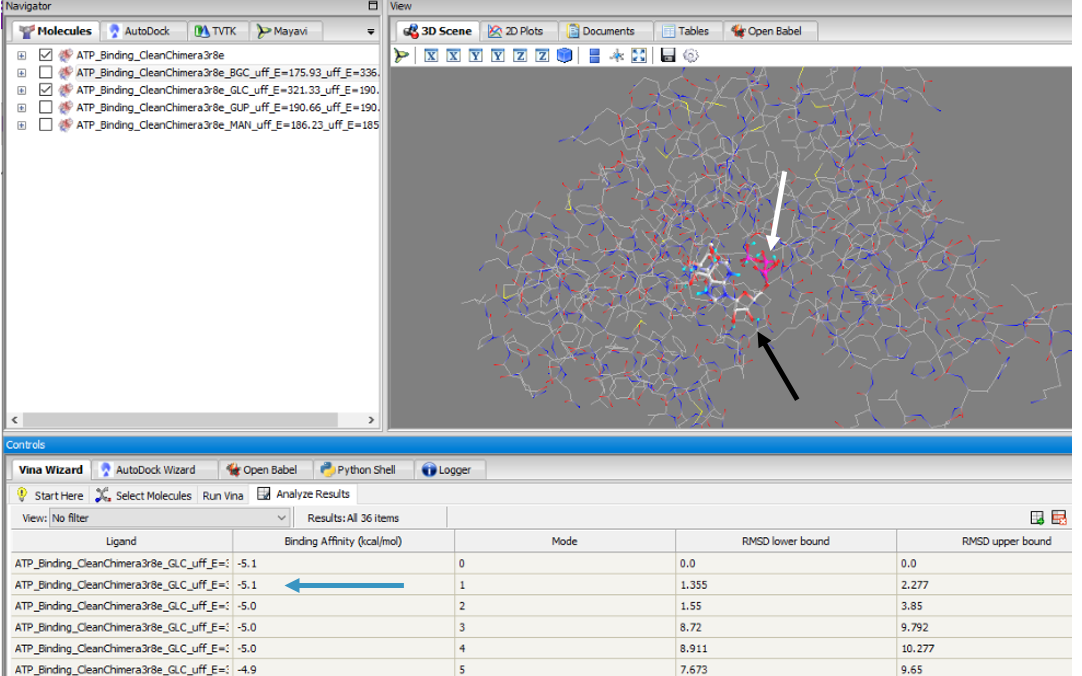
These results along with supporting evidence from the other in silico tools allowed us to then conduct in silico docking experiments to further confirm our hypothesis. Below is a figure with the PyRx docking results of glucose and ATP with our protein. | These results along with supporting evidence from the other in silico tools allowed us to then conduct in silico docking experiments to further confirm our hypothesis. Below is a figure with the PyRx docking results of glucose and ATP with our protein. | ||
| - | [[Image: | + | [[Image:PyRx1.png]] |
The -5.1 kcal/mol value shows that our protein of interest hypothesis of a glucose kinase is strong. The confidence behind our in silico results allowed us to move into testing our hypothesis in vitro. | The -5.1 kcal/mol value shows that our protein of interest hypothesis of a glucose kinase is strong. The confidence behind our in silico results allowed us to move into testing our hypothesis in vitro. | ||
== Experimental Results/Function == | == Experimental Results/Function == | ||
Revision as of 01:01, 21 April 2022
Characterization of the 3r8e Protein, a Novel Gluco Kinase
| |||||||||||
References
1. Blastp [Internet]. Bethesda (MD): Natiobal Library of Medicine (US), National Center for Biotechnology Information; 2004- [cited 2022 March]. Available from: (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)
2. BASIL. https://basilbiochem.github.io/basil/
3. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
4. Small- Molecule Library Screening by Docking with PyRx. .Dallakyan S, Olson AJ Methods Mol Biol. 2015;1263:243-50. The full-text is available at https://www.researchgate.net/publications/2739554875. Small-Molecule Library Screening by Docking with PyRx.
5. Pfam: The Protein families database in 2021 J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
6. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky