BASIL2022GV3R8E
From Proteopedia
| Line 10: | Line 10: | ||
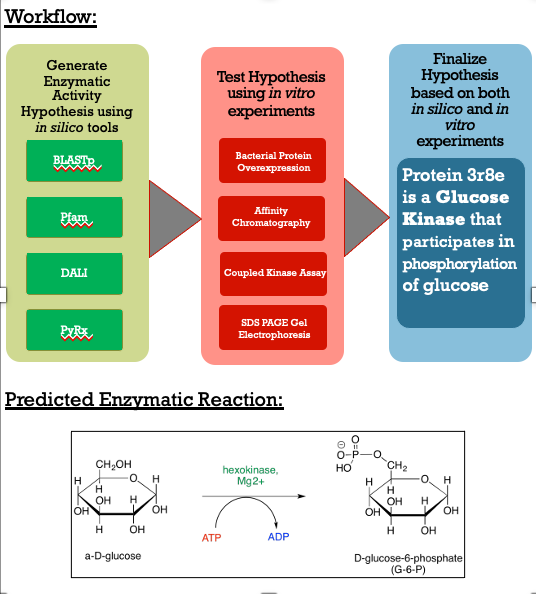
As you can see in the workflow portion above, we used a variety of in silico tools such as BLASTp, Pfam, DALI, PyRx, and PyMol to help us generate a hypothesis for our uncharacterized proteins function. Using the FASTA sequence found in the Protein Data Bank file, we then were able to find similarities between 3r8e and other characterized proteins. While exploring the DALI database, a significant structural alignment hit was found with protein 3vov. Alignment and highly conserved amino acids can be seen below. | As you can see in the workflow portion above, we used a variety of in silico tools such as BLASTp, Pfam, DALI, PyRx, and PyMol to help us generate a hypothesis for our uncharacterized proteins function. Using the FASTA sequence found in the Protein Data Bank file, we then were able to find similarities between 3r8e and other characterized proteins. While exploring the DALI database, a significant structural alignment hit was found with protein 3vov. Alignment and highly conserved amino acids can be seen below. | ||
| - | [[Image: | + | [[Image:3R8EDALI.png|300px|left|thumb|]] |
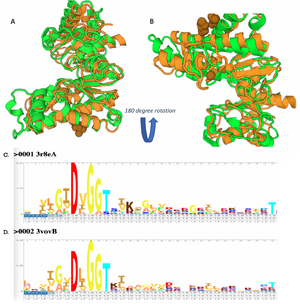
These results along with supporting evidence from the other in silico tools allowed us to then conduct in silico docking experiments to further confirm our hypothesis. Below is a figure with the PyRx docking results of glucose and ATP with our protein. | These results along with supporting evidence from the other in silico tools allowed us to then conduct in silico docking experiments to further confirm our hypothesis. Below is a figure with the PyRx docking results of glucose and ATP with our protein. | ||
| - | [[Image:PyRx1.png]] | + | [[Image:PyRx1.png|300px|left|thumb|]] |
The -5.1 kcal/mol value shows that our protein of interest hypothesis of a glucose kinase is strong. The confidence behind our in silico results allowed us to move into testing our hypothesis in vitro. Because ATP is a commonality amongst sugar kinases, an <scene name='90/904995/3r8ec_w_glc_and_atp/1'>interactive structure</scene> has been provided to represent glucose and ATP in the active site. | The -5.1 kcal/mol value shows that our protein of interest hypothesis of a glucose kinase is strong. The confidence behind our in silico results allowed us to move into testing our hypothesis in vitro. Because ATP is a commonality amongst sugar kinases, an <scene name='90/904995/3r8ec_w_glc_and_atp/1'>interactive structure</scene> has been provided to represent glucose and ATP in the active site. | ||
== Experimental Results/Function == | == Experimental Results/Function == | ||
| Line 19: | Line 19: | ||
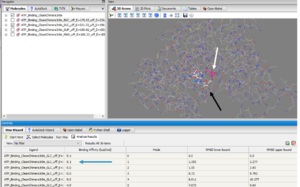
Below are the results of our Uncoupled Kinase Assay. Our results from this assay further supports our idea of protein 3r8e assisting in the phosphorylation of glucose. | Below are the results of our Uncoupled Kinase Assay. Our results from this assay further supports our idea of protein 3r8e assisting in the phosphorylation of glucose. | ||
| - | [[Image:KinaseAssayResults.png]] | + | [[Image:KinaseAssayResults.png|300px|left|thumb|]] |
== Project Implications == | == Project Implications == | ||
The goal of this project is to explore the methods it takes to characterize a putative kinase. To do this, we became familiar with online alignment, structure, and function tools, paired with a variety of in vitro lab experiments, such as bacterial protein overexpression, affinity chromatography, coupled kinase assays, and SDS PAGE Gel Electrophoresis. Proteins are biomolecules essential to organisms' survival and understanding how they work in result of their function is pivotal for advances in modern medicine, scientific research, and agriculture. | The goal of this project is to explore the methods it takes to characterize a putative kinase. To do this, we became familiar with online alignment, structure, and function tools, paired with a variety of in vitro lab experiments, such as bacterial protein overexpression, affinity chromatography, coupled kinase assays, and SDS PAGE Gel Electrophoresis. Proteins are biomolecules essential to organisms' survival and understanding how they work in result of their function is pivotal for advances in modern medicine, scientific research, and agriculture. | ||
Revision as of 03:04, 26 April 2022
Characterization of the 3r8e Protein, a Novel Gluco Kinase
| |||||||||||
References
1. Blastp [Internet]. Bethesda (MD): Natiobal Library of Medicine (US), National Center for Biotechnology Information; 2004- [cited 2022 March]. Available from: (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)
2. BASIL. https://basilbiochem.github.io/basil/
3. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
4. Small- Molecule Library Screening by Docking with PyRx. .Dallakyan S, Olson AJ Methods Mol Biol. 2015;1263:243-50. The full-text is available at https://www.researchgate.net/publications/2739554875. Small-Molecule Library Screening by Docking with PyRx.
5. Pfam: The Protein families database in 2021 J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
6. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky