Abstract
The Protein Data Bank (PDB) contains approximately 188 thousand protein structures, 5000 of which have not been assigned a specific function. As a part of the Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) project, we were tasked with analyzing and determining the function of one of these proteins, PDB ID 3r8e. This protein is a putative kinase, which is of interest due to the key roles kinases play in many cellular processes. Utilizing the modules the BASIL consortium provides, a series of in silico and in vitro experiments were conducted. The 3r8e protein was first studied using a variety of in silico tools, including BLASTp, Pfam, and DALI. Based on our in silico results, glucose was determined to be the most likely substrate for 3r8e and was used for further in vitro characterization of the protein. To confirm the in silico function prediction for the 3r8e protein, bacterial protein overexpression, affinity chromatography purification, and coupled kinase activity assays were utilized. Multiple sugar substrates for 3r8e were tested, including glucose. The coupled kinase assay results confirmed that 3r8e likely plays a role in glucose phosphorylation, aligning with our in silico conclusions. Future directions include repeating the above experiments and testing additional hexose substrates to further characterize the enzymatic activity of 3r8e. Overall, we have strong preliminary evidence that the 3r8e protein is a glucose kinase and future work will allow us to confirm this.
Introduction
Like many proteins with solved crystal structures, protein 3r8e has an uncharacterized and unconfirmed function. Besides the relationship with bacteria in soil, the role of this protein is still undetermined. As apart of a research project under the Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) consortium, our group was tasked with characterizing and identifying the function of this protein to potentially provide further insight of the protein's relationship to the bacteria. Below is the general workflow of how we got our results.
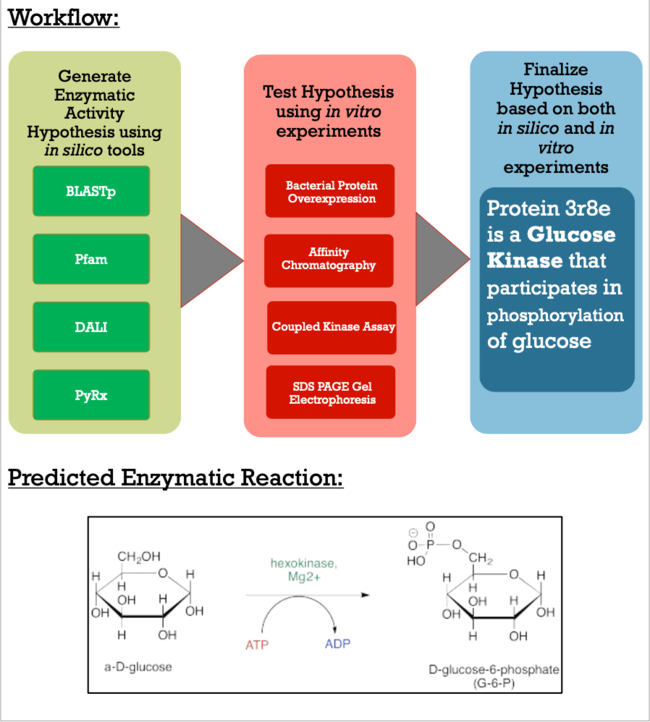
Methods
As you can see in the workflow portion above, we used a variety of in silico tools such as BLASTp, Pfam, DALI, PyRx, and PyMol to help us generate a hypothesis for our uncharacterized proteins function. Using the FASTA sequence found in the Protein Data Bank file, we then were able to find similarities between 3r8e and other characterized proteins. While exploring the DALI database, a significant structural alignment hit was found with protein 3vov. Alignment and highly conserved amino acids can be seen below.
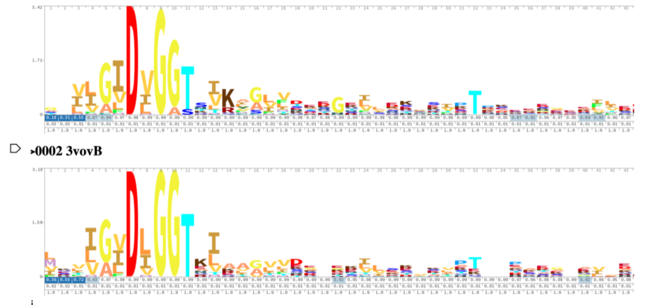
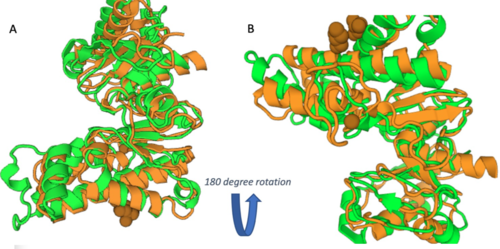
These results, along with supporting evidence from the previous in silico tools, allowed us to continue testing our hypothesis. To begin, we visualized our hypothesized substrate within the active site using mesh view in PyMol. This PyMol provided below visualization allowed us to see how involved glucose is in our active site. The interactions between glucose (yellow), ATP (blue), and the protein help solidify our initial thoughts.
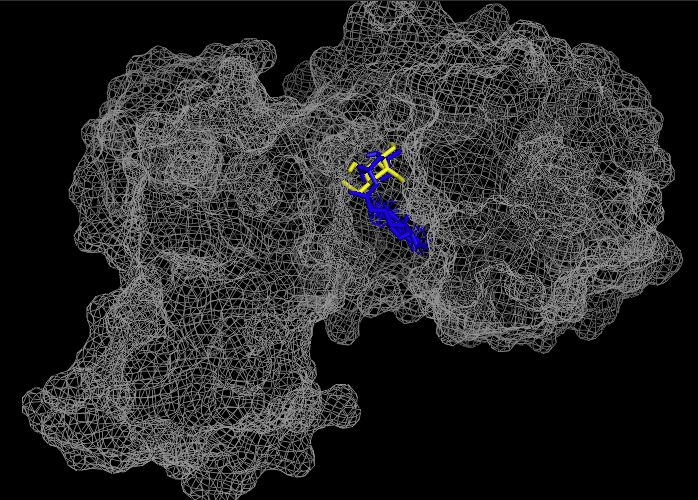
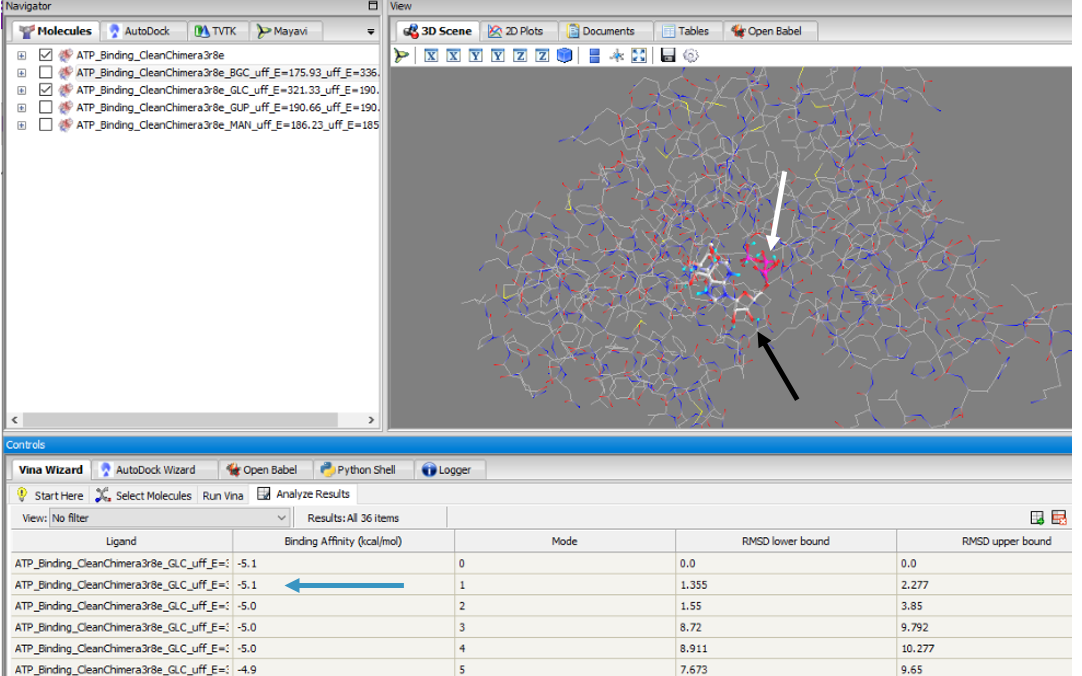
To validate that glucose actually binds and interacts with our protein of interest, we conducted a PyRx in silico docking experiment with a total of five hexose substrates. Below is the figure with the PyRx docking results of glucose and ATP with our protein. Other substrates tested include fructose, galactose, lactose, and ribose, however, experimental in silico docking results for those substrates were significantly less than glucose. The binding affinity of glucose (-5.1 kcal/mol) strengthens our idea that glucose is phosphorylated by our protein of interest. The confidence behind our in silico results allowed us to move into testing our hypothesis in vitro and because ATP is a commonality amongst sugar kinases, an has been provided to represent interactions of glucose and ATP in the active site.
Experimental Results/Function
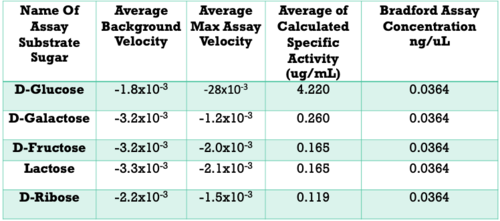
Below are the results of our Uncoupled Kinase Assay. Our results from this assay further supports our idea of protein 3r8e assisting in the phosphorylation of glucose. A total of five hexose substrates were tested in vitro, detailed in the table below. Based on these results, we were able to strengthen our initial hypothesis and continue characterization.
For further validation, we conducted an SDS analysis and provided below is the gel image. Results were not as clear as anticipated, and in future studies, we would need to utilize different chromatography methods to yield higher quality protein concentrations and conduct a pre and post induction to visualize the purity of our protein.
Project Implications
The goal of this project is to explore the methods it takes to characterize a putative kinase. To do this, we became familiar with online alignment, structure, and function tools, paired with a variety of in vitro lab experiments, such as bacterial protein overexpression, affinity chromatography, coupled kinase assays, and SDS PAGE Gel Electrophoresis. Proteins are biomolecules essential to organisms' survival and understanding how they work in result of their function is pivotal for advances in modern medicine, scientific research, and agriculture.
Conclusions/Future Direction
Conclusions:
- D-Glucose exhibited a specific activity of 4.22 ug/mL
- Specific activity: D-Glucose >> D-Galactose > D-Fructose > Lactose > D-Ribose
- Protein 3r8e is a Glucose Kinase
- Glucose is phosphorylated by our protein
Future Direction:
- Verify experimental results
- Publish results for further application
- Authenticate protein function using further kinase characterization protocols






