Sandbox Reserved 1741
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== Function == | == Function == | ||
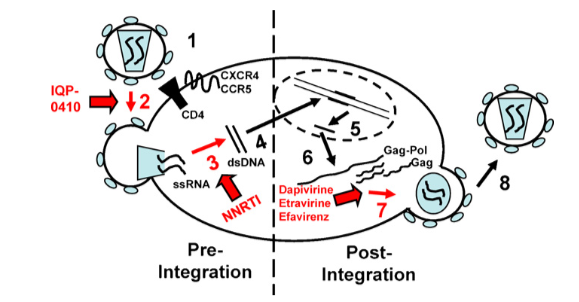
| - | HIV is a retrovirus, which means that it only carries single stranded RNA, and so relies on reverse transcription to propagate itself to new cells (2). HIV reverse transcriptase (RT) is a heterodimer consisting of a p51 and a p66 subunit, the latter of which contains catalytically active DNA polymerase and RNase H domains, essentially functioning as a catalyst for the reverse transcription reaction (3). The enzyme RT is responsible for turning the infectious single-stranded ssRNA genome into double-stranded dsDNA provirus to integrate the virus into a new host cell (4). HIV integrase then integrates it into the host chromosome. The retro transcription process begins when the HIV particle fuses with the membrane of the host due to interactions between the envelope of the glycoprotein and coreceptors on the surface of the host cell (5). The infectious content of the HIV particle is then released into the cytoplasm of the host cell. Here the ssRNA serves as a template for HIV RT to form the dsDNA provirus. The dsDNA is then imported into the nucleus, which is then integrated into the host chromosome by another enzyme, HIV integrase. Once the dsDNA is integrated into the nucleus it makes modified mRNA that code for viral proteins and new ssRNA gnomes that combine to form new viral HIV particles (4). Some viral proteins that are made by the modified mRNA include Gag, Gag-Pol, and Env. The modified mRNA also codes for the new accessory proteins Nef, Vif, Vpr, and Vpu. (6) These viral proteins then form chains, which combine with the ssRNA to form new viral particles. These particles then release (bud) from the cell, taking some of the cell membrane with it. (6) The viral particles seek out new host cells to infect, spreading the virus through the system and starting the process anew. Figure #1 shows a graphical representation of the HIV life cycle. | + | HIV is a retrovirus, which means that it only carries single stranded RNA, and so relies on reverse transcription to propagate itself to new cells (2). '''HIV reverse transcriptase''' (RT) is a heterodimer consisting of a p51 and a p66 subunit, the latter of which contains catalytically active DNA polymerase and RNase H domains, essentially functioning as a catalyst for the reverse transcription reaction (3). The enzyme RT is responsible for turning the infectious single-stranded '''ssRNA''' genome into double-stranded '''dsDNA''' provirus to integrate the virus into a new host cell (4). HIV integrase then integrates it into the host chromosome. The retro transcription process begins when the HIV particle fuses with the membrane of the host due to interactions between the envelope of the glycoprotein and coreceptors on the surface of the host cell (5). The infectious content of the HIV particle is then released into the cytoplasm of the host cell. Here the ssRNA serves as a template for HIV RT to form the dsDNA provirus. The dsDNA is then imported into the nucleus, which is then integrated into the host chromosome by another enzyme, HIV integrase. Once the dsDNA is integrated into the nucleus it makes modified '''mRNA''' that code for viral proteins and new ssRNA gnomes that combine to form new viral HIV particles (4). Some viral proteins that are made by the modified mRNA include '''Gag''', '''Gag-Pol''', and '''Env'''. The modified mRNA also codes for the new accessory proteins '''Nef''', '''Vif''', '''Vpr''', and '''Vpu'''. (6) These viral proteins then form chains, which combine with the ssRNA to form new viral particles. These particles then release (bud) from the cell, taking some of the cell membrane with it. (6) The viral particles seek out new host cells to infect, spreading the virus through the system and starting the process anew. Figure #1 shows a graphical representation of the HIV life cycle. |
| Line 12: | Line 12: | ||
Figure #1 (6) | Figure #1 (6) | ||
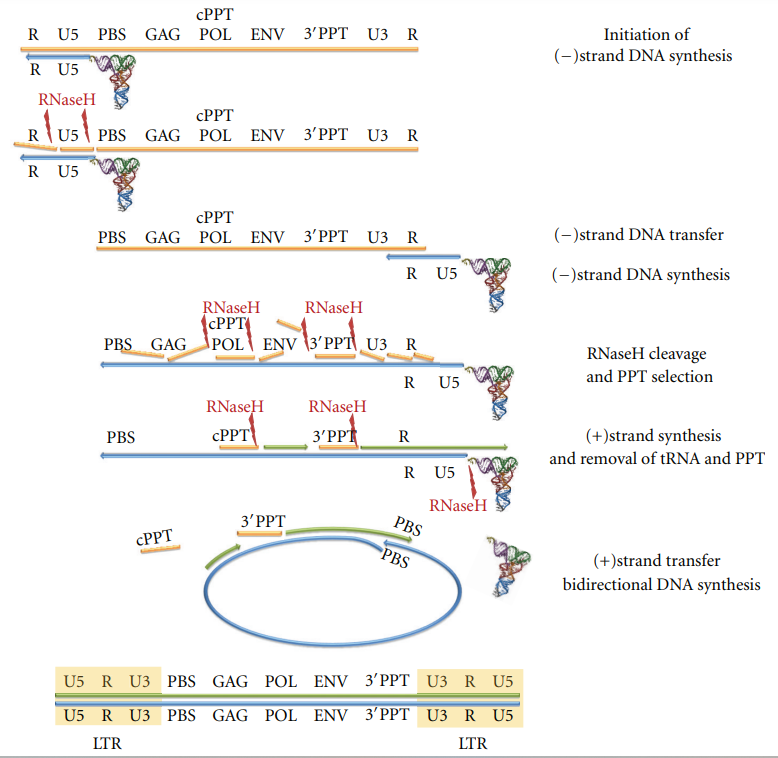
| - | The actual process of HIV reverse transcription begins when the host cell tRNALsy3 hybridizes at the 5’-end of the (+) strand on the RNA genome. tRNALys3 is then used as a primer for the (-) strand DNA synthesis. DNA synthesis continues until the 5’ end of the RNA strand. The next step in the process involves RNase hydrolysis of the RNA/DNA hybrid to determine the (-) strand strong stop DNA by exposing the ssDNA product. The (-) strand DNA is then lengthened further and hybridized with the R region at the 3’-end of the ssRNA strand to transfer it. DNA synthesis continues when the RNase H function cleaves the RNA strand in the RNA/DNA hybrid at multiple different points, leaving two specific sections undamaged. These sections are cPPT and 3’PPt, both of which are resistant to the RNase H cleavage. DNA synthesis for the (-) strand is initiated again, only this time using the PPT strand fragments as primers. Hydrolysis of the PPT segments and the junction of the tRNA/DNA hybrid is then initiated by the RNase H, which frees up the PBS sequence of the (+) strand DNA. The PBS sequence from the (+) strand of DNA then anneals to the PBS on the (-) strand DNA. The DNA synthesis then continues, using strand displacement synthesis to get a linear dsDNA with long terminal repeats on both ends as the product (4). Figure #2 shows a graphical representation of reverse transcription. | + | The actual process of HIV reverse transcription begins when the host cell '''tRNALsy3''' hybridizes at the 5’-end of the (+) strand on the RNA genome. tRNALys3 is then used as a primer for the (-) strand DNA synthesis. DNA synthesis continues until the 5’ end of the RNA strand. The next step in the process involves '''RNase hydrolysis''' of the RNA/DNA hybrid to determine the (-) strand strong stop DNA by exposing the ssDNA product. The (-) strand DNA is then lengthened further and hybridized with the R region at the 3’-end of the ssRNA strand to transfer it. DNA synthesis continues when the '''RNase H''' function cleaves the RNA strand in the RNA/DNA hybrid at multiple different points, leaving two specific sections undamaged. These sections are '''cPPT''' and '''3’PPt''', both of which are resistant to the RNase H cleavage. DNA synthesis for the (-) strand is initiated again, only this time using the PPT strand fragments as primers. Hydrolysis of the PPT segments and the junction of the tRNA/DNA hybrid is then initiated by the RNase H, which frees up the '''PBS sequence''' of the (+) strand DNA. The PBS sequence from the (+) strand of DNA then anneals to the PBS on the (-) strand DNA. The DNA synthesis then continues, using '''strand displacement synthesis''' to get a linear dsDNA with long terminal repeats on both ends as the product (4). Figure #2 shows a graphical representation of reverse transcription. |
Current revision
| This Sandbox is Reserved from August 30, 2022 through May 31, 2023 for use in the course Biochemistry I taught by Kimberly Lane at the Radford University, Radford, VA, USA. This reservation includes Sandbox Reserved 1730 through Sandbox Reserved 1749. |
To get started:
More help: Help:Editing |
| |||||||||||