We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1791
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | {{Template:CH462_Biochemistry_II_2023}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | |
| - | + | ||
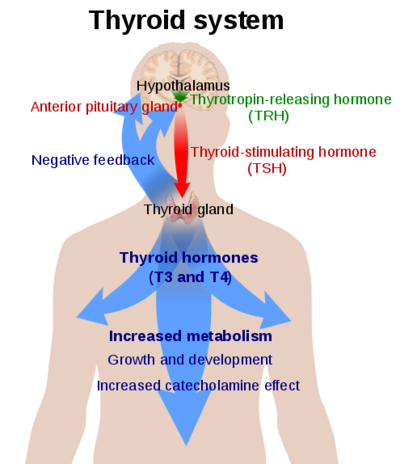
==Thyrotropin Receptor== | ==Thyrotropin Receptor== | ||
<StructureSection load='7xw5' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='7xw5' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| Line 23: | Line 22: | ||
=== Binding Pocket=== | === Binding Pocket=== | ||
<scene name='95/952719/Binding_pocket/2'>Binding Pocket</scene> | <scene name='95/952719/Binding_pocket/2'>Binding Pocket</scene> | ||
| + | *a concave structure with many polar residues | ||
== Active vs. Inactive State == | == Active vs. Inactive State == | ||
| - | *<scene name='95/952719/Active_form/2'>Active Form</scene> | + | *The active form of TSHR is found when bound by TSH or M22. The structure can be seen as straight. <scene name='95/952719/Active_form/2'>Active Form</scene> |
| - | *<scene name='95/952719/Inactive_form/2'>Inactive form</scene> | + | *The inactive form of TSHR is found when bound with K1. The overall structure of the molecule when K1 is bound is bent. <scene name='95/952719/Inactive_form/2'>Inactive form</scene> |
| + | |||
== Specific Residues == | == Specific Residues == | ||
| + | *LYS 58 interacts with Glu 118 on each of the antibodies | ||
| + | *LYS 209 interacts with Asp 11 on each of the antibodies | ||
| + | *When in the inactive form, Lys 209 does not interact with any residue but Lys 58 has interaction with Glu 118 and this interaction pulls the molecule into the bent position. | ||
*<scene name='95/952719/Specific_residues/1'>LYS Residues</scene> | *<scene name='95/952719/Specific_residues/1'>LYS Residues</scene> | ||
*<scene name='95/952719/Specific_residues/2'>LYS closer look</scene> | *<scene name='95/952719/Specific_residues/2'>LYS closer look</scene> | ||
Revision as of 17:40, 24 March 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
Thyrotropin Receptor
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Student Contributions
- Alex Kem
- Grace Lane