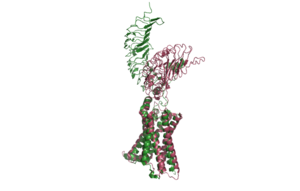We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1792
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
== Active vs Inactive State== | == Active vs Inactive State== | ||
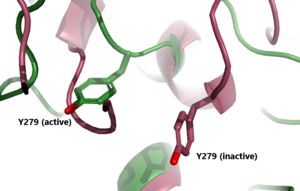
[[Image:Inactive v active proteopedia.png|300 px|right|thumb| Figure 2: An overview of the Inactive (pink) vs Active (green) state of TSHR. PDB: 7WX5]] [[Image:Inactive v active residue.png|300 px|right|thumb| Figure 3: A zoomed in view of the Y279 residue in the Hinge Region of TSHR, showing the 6 angstrom move of Y279 during the activation of TSHR. Active TSHR is shown in green (PDB: 7t9i) and inactive TSHR is shown in pink (PDB: 7t9m).]] | [[Image:Inactive v active proteopedia.png|300 px|right|thumb| Figure 2: An overview of the Inactive (pink) vs Active (green) state of TSHR. PDB: 7WX5]] [[Image:Inactive v active residue.png|300 px|right|thumb| Figure 3: A zoomed in view of the Y279 residue in the Hinge Region of TSHR, showing the 6 angstrom move of Y279 during the activation of TSHR. Active TSHR is shown in green (PDB: 7t9i) and inactive TSHR is shown in pink (PDB: 7t9m).]] | ||
| - | When TSHR is not bound to TSH, it is in the <scene name='95/952720/Inactivetshr/ | + | When TSHR is not bound to TSH, it is in the <scene name='95/952720/Inactivetshr/8'>inactive state</scene>. This is also considered the "down" state because the LRRD is pointing down. When TSH binds to TSHR, steric clashing between TSH and the cell-membrane cause TSHR to take on the <scene name='95/952720/Inactivetshr/6'>active or "up" state</scene> (fig 2). During this transition, the Extracellular domains rotate 55° along an axis. This rotation is caused by conformational changes within the <scene name='95/952720/Hinge_region_spin/1'>Hinge Region</scene>, specifically at the <scene name='95/952720/Hinge_region_residues/2'>Y279 residue</scene>. This residue moves 6 angstroms relative to I486, which is a residue located in the Transmembrane Region <ref name="Faust"/> (Fig 3). |
== Specific Residues == | == Specific Residues == | ||
Revision as of 16:41, 7 April 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
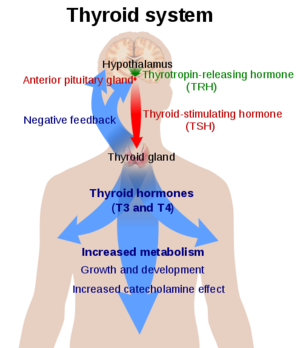
Thyroid Stimulating Hormone Receptor (TSHR)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 3.2 Faust B, Billesbolle CB, Suomivuori CM, Singh I, Zhang K, Hoppe N, Pinto AFM, Diedrich JK, Muftuoglu Y, Szkudlinski MW, Saghatelian A, Dror RO, Cheng Y, Manglik A. Autoantibody mimicry of hormone action at the thyrotropin receptor. Nature. 2022 Aug 8. pii: 10.1038/s41586-022-05159-1. doi:, 10.1038/s41586-022-05159-1. PMID:35940205 doi:http://dx.doi.org/10.1038/s41586-022-05159-1
- ↑ Duan J, Xu P, Luan X, Ji Y, He X, Song N, Yuan Q, Jin Y, Cheng X, Jiang H, Zheng J, Zhang S, Jiang Y, Xu HE. Hormone- and antibody-mediated activation of the thyrotropin receptor. Nature. 2022 Aug 8. pii: 10.1038/s41586-022-05173-3. doi:, 10.1038/s41586-022-05173-3. PMID:35940204 doi:http://dx.doi.org/10.1038/s41586-022-05173-3
Student Contributors
- Alex Kem
- Grace Lane