This is a default text for your page BASIL2023GVP30646. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Abstract
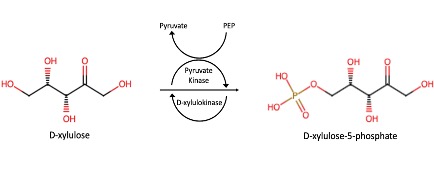
The Protein Data Bank (PDB) and UniProt contain a wealth of protein structures, many of which do not have a known function. One such protein is P30646, an uncharacterized sugar kinase. Sugar kinases play a role in a variety of metabolic processes within the body, so understanding their functions is of great importance. Our work aimed to further characterize P30646 through the identification of a substrate for this enzyme. Utilizing materials Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) project, our group combined in silico and in vitro data for a full analysis of P30646. Computation tools including BLAST, DALI, SPRITE, and InterPro allowed our group to compare the structural and sequential similarities between P30646 and proteins with known functions. In silico analysis indicated that P30646 had xylulokinase activity, and xylulose was tested as a substrate in vitro using a coupled kinase assay. The kinase assay indicated that P30646 does have strong enzymatic activity for xylulose. The combination of our in silico and in vitro results provide a strong indication that P30646 is a sugar kinase with affinity for xylulose breakdown.
Background
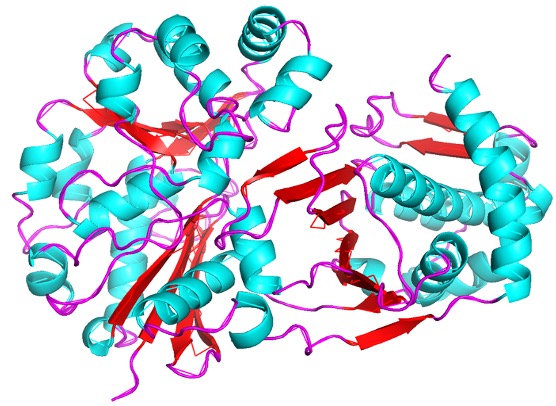
The study of protein structure and function is essential, as the structure of a protein is often closely related to its function. This project investigated the protein P30646 (UniProt), a protein with a known structure but an unknown function. It is understood that P30646 is an uncharacterized sugar kinase that is involved in the metabolic processes of Caenorhabditis elegans. To determine its function as well as a potential substrate, we first conducted an in silico exploration. These tools included DALI, InterPro, PredictProtein, and BLASTp. Each of these tools draws upon data for proteins with known functions to generate hypotheses about the function of the protein of interest. We then tested these hypotheses in vitro by overexpressing and purifying P30646, and then analyzing the protein through a Bradford assay, a coupled kinase assay, and SDS-PAGE. Through the analysis of the results obtained from these computational and laboratory methods, we aimed to identify the function of P30646 and gain insight into its role in the metabolism of C. elegans
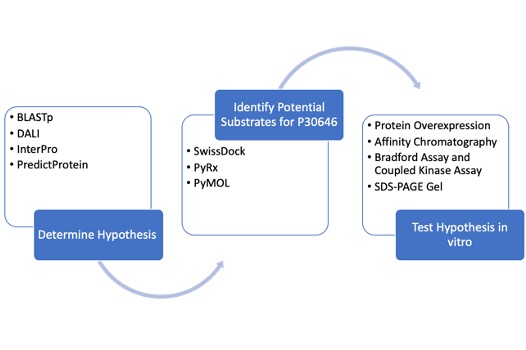
Workflow
Predicted Mechanism
P30646 Structure
Methods
Experimental Results
Conclusions
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.



