BASIL2023GVP30646
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Background == | == Background == | ||
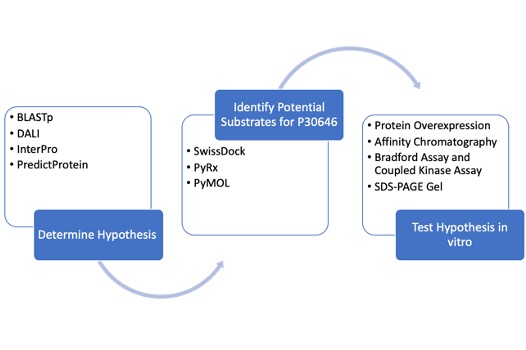
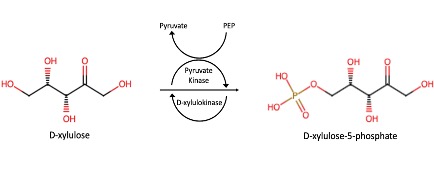
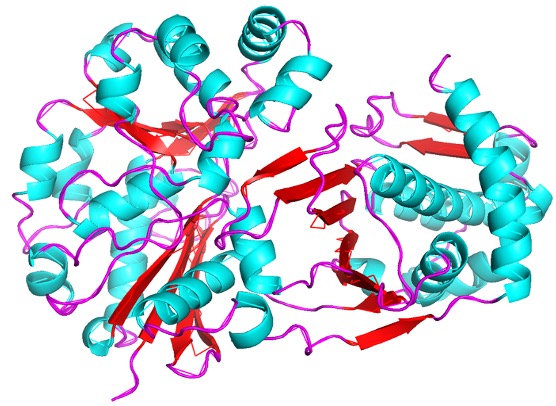
| - | The study of protein structure and function is essential, as the structure of a protein is often closely related to its function. This project investigated the protein P30646 (UniProt), a protein with a known structure but an unknown function. It is understood that P30646 is an uncharacterized sugar kinase that is involved in the metabolic processes of Caenorhabditis elegans | + | The study of protein structure and function is essential, as the structure of a protein is often closely related to its function. This project investigated the protein P30646 (UniProt), a protein with a known structure but an unknown function. It is understood that P30646 is an uncharacterized sugar kinase that is involved in the metabolic processes of Caenorhabditis elegans. Through the analysis of results obtained through computational and laboratory methods, this work aims to identify the function of P30646 and gain insight into its role in the metabolism of C. elegans. |
Revision as of 23:27, 11 April 2023
Characterization of Novel Xylulokinase P30646
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644