User:Eric Martz/AlphaFold3 case studies
From Proteopedia
(→Q381Q7: AlphaFold3 vs AlphaFold2 Database) |
(→Q381Q7: AlphaFold3 vs AlphaFold2 Database) |
||
| Line 22: | Line 22: | ||
===Q381Q7: AlphaFold3 vs AlphaFold2 Database=== | ===Q381Q7: AlphaFold3 vs AlphaFold2 Database=== | ||
<table class="wikitable"><tr><td width="400" rowspan="2"> | <table class="wikitable"><tr><td width="400" rowspan="2"> | ||
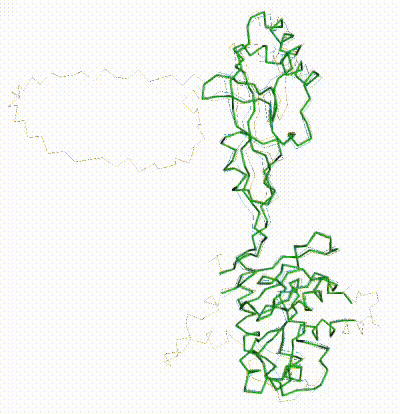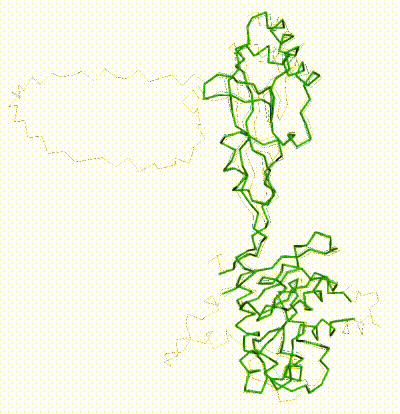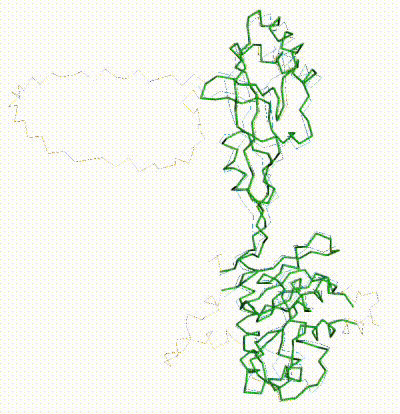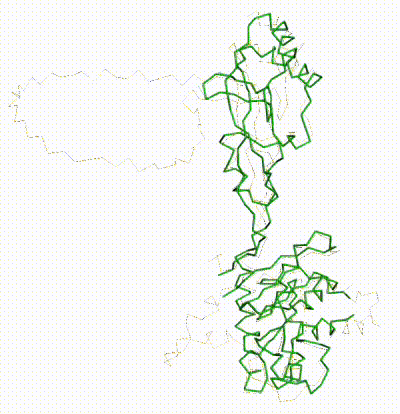
| - | + | The long disordered loop was in different positions in the AlphaFold3 vs. AlphaFold2 Database models. When it was included, FATCAT superposed it between models, causing the RMSD to be 4.8-4.9 Å (rigid or flexible superposition). | |
| + | |||
| + | Deleting that loop from the models resulted in a much better flexible superposition RMSD value of '''2.0 Å''' for 312 alpha carbons (98% of the 317 present). This close superposition required flexibility. The rigid FATCAT superposition still had RMSD 4.5 Å. The morph shows bending and twisting between the two models. | ||
</td><td width="262"> | </td><td width="262"> | ||
[[Image:Q381q7-af3-vs-af2.gif]] | [[Image:Q381q7-af3-vs-af2.gif]] | ||
Revision as of 22:45, 12 November 2024
The following case studies of predictions by AlphaFold3 were done in November 2024. Superpositions and RMSD values were obtained from FATCAT. Animations were captured from FATCAT.
Contents |
AlphaFold3 Example 8AW3
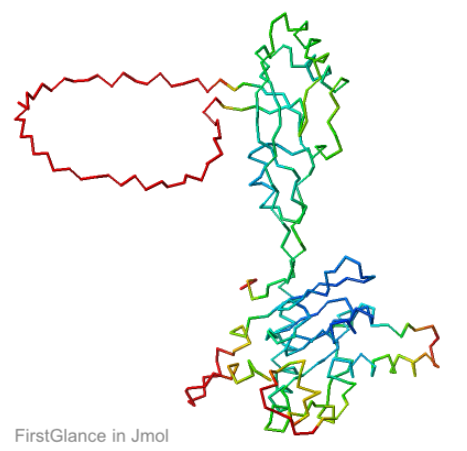
Q381Q7 (tRNA A34 deaminase) is the longest chain in the example 8aw3 provided by the AlphaFold3 Server. In 8aw3, its chain name is "3". 8aw3 is a cryo-EM structure with resolution 3.6 Å. 8aw3 indicates that the experimental material included full-length chain 3 of 369 residues, but 28 end-residues lack coordinates, so the coordinates run from UniProt 4-344, length 341. Within this range, there are two loops lacking coordinates of lengths 49 and 13, leaving 279 amino acids with coordinates. Neither missing loop is predicted to be disordered at UniProt, although the disorder estimate is high for both.
Q381Q7: AlphaFold3 vs Empirical Structure
The overall 4-chain prediction (for the complex in 8aw3) has pTM of 0.75, at the low end of the range 0.7-0.9 deemed "confident" by the server. Both of the missing loops have "no-confidence" (red) coordinates. The longer one is modeled as a large circle protruding from the surface of the protein, typical for loops with intrinsic disorder. The average pLDDT for chain 3 is 68 (weak confidence), but with the large disordered loop removed, this increases to 74[1].
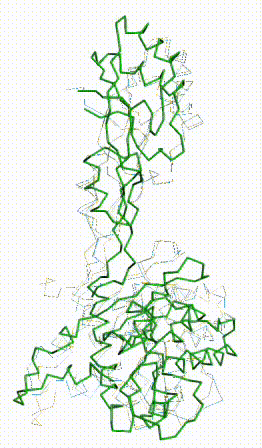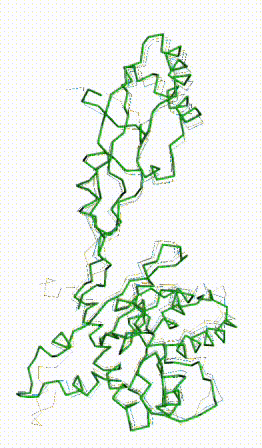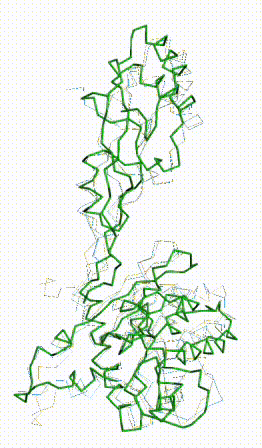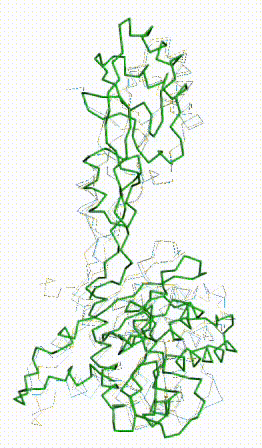
The AlphaFold3 prediction superposes (FATCAT rigid) with all 279 alpha carbons of chain 3 of 8aw3 with RMSD 2.2 Å. The morph shows the largest discrepancy in the red (no confidence) loop at center bottom, which has sequence SNSGCRKSNR (238-247, UniProt = 8aw3). These residues have coordinates in 8aw3, but 3 residues, CRK, have incomplete sidechains.
|
AlphaFold3 prediction for Q38107. |
FATCAT morph between AlphaFold3 prediction and chain 3 of 8aw3. |
Q381Q7: AlphaFold3 vs AlphaFold2 Database
|
The long disordered loop was in different positions in the AlphaFold3 vs. AlphaFold2 Database models. When it was included, FATCAT superposed it between models, causing the RMSD to be 4.8-4.9 Å (rigid or flexible superposition). Deleting that loop from the models resulted in a much better flexible superposition RMSD value of 2.0 Å for 312 alpha carbons (98% of the 317 present). This close superposition required flexibility. The rigid FATCAT superposition still had RMSD 4.5 Å. The morph shows bending and twisting between the two models. | |
|
FATCAT morph between AlphaFold3 prediction and AlphaFold2 Database prediction for Q381Q7. |
Notes
- ↑ Average pLDDT values were obtained with FirstGlance in Jmol, which reports the minimum, average, and maximum in the upper right as "Reliability".