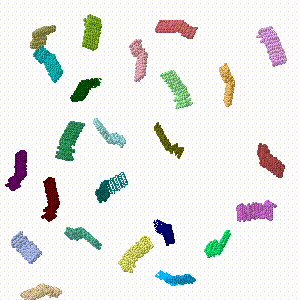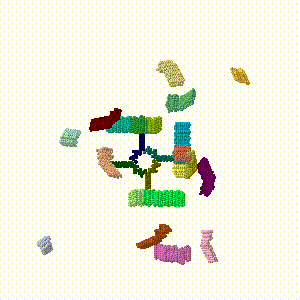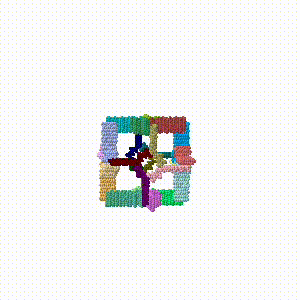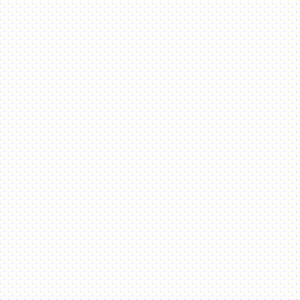Synthetic nanomaterials from standardized protein blocks
From Proteopedia
(Difference between revisions)
| Line 112: | Line 112: | ||
</StructureSection> | </StructureSection> | ||
| + | |||
==Method of simulating assembly== | ==Method of simulating assembly== | ||
| - | The cage model (Cube with no vertices [[Image:Huddy2024-cube-noverts-cage-o4-32.pdb.gz]]) has about 272,000 atoms including hydrogens. It was simplified to alpha carbon atoms only (17,256) using the [[Jmol/Application|Jmol.jar Java application]]. The assembled cage has 24 protein chains. In Jmol.jar, 4 chains were dragged out of the assembly to the periphery of the viewport, and each was rotated arbitrarily. | + | The cage model (Cube with no vertices [[Image:Huddy2024-cube-noverts-cage-o4-32.pdb.gz]]) has about 272,000 atoms including hydrogens. It was simplified to alpha carbon atoms only (17,256) using the [[Jmol/Application|Jmol.jar Java application]] using Jmol commands "select *.ca; write 0.pdb;". The assembled cage has 24 protein chains. |
| + | |||
| + | #In Jmol.jar, 4 chains were dragged out of the assembly to the periphery of the viewport, and each was rotated arbitrarily using the Jmol setting "set picking dragmolecule" (holding down Alt enables rotation). The resulting state was written into a PDB file. | ||
| + | |||
| + | #Repeating this drag/rotate save process six times produced a final model in which none of the protein chains were in their original assembled positions. This resulted in six PDB files 1.pdb, ... 6.pdb. | ||
| + | |||
| + | # The [https://www.bioinformatics.org/pdbtools/morph2 linear morph server by Karsten Theis] was used on each pair of PDB files to morph in reverse (towards assembly), producing six morphs 6->5, 5->4, ... 1->0 (where 0 is the fully assembled cube). | ||
| + | |||
==See Also== | ==See Also== | ||
*Related work from the Baker group includes <i>Bond-centric modular design of protein assemblies</i> by Wang <i>et al.</i>, 2024<ref name="wang">PMID: 39416012</ref>. | *Related work from the Baker group includes <i>Bond-centric modular design of protein assemblies</i> by Wang <i>et al.</i>, 2024<ref name="wang">PMID: 39416012</ref>. | ||
Revision as of 18:11, 10 February 2025
| |||||||||||
Method of simulating assembly
The cage model (Cube with no vertices Image:Huddy2024-cube-noverts-cage-o4-32.pdb.gz) has about 272,000 atoms including hydrogens. It was simplified to alpha carbon atoms only (17,256) using the Jmol.jar Java application using Jmol commands "select *.ca; write 0.pdb;". The assembled cage has 24 protein chains.
- In Jmol.jar, 4 chains were dragged out of the assembly to the periphery of the viewport, and each was rotated arbitrarily using the Jmol setting "set picking dragmolecule" (holding down Alt enables rotation). The resulting state was written into a PDB file.
- Repeating this drag/rotate save process six times produced a final model in which none of the protein chains were in their original assembled positions. This resulted in six PDB files 1.pdb, ... 6.pdb.
- The linear morph server by Karsten Theis was used on each pair of PDB files to morph in reverse (towards assembly), producing six morphs 6->5, 5->4, ... 1->0 (where 0 is the fully assembled cube).
See Also
- Related work from the Baker group includes Bond-centric modular design of protein assemblies by Wang et al., 2024[9].
- Metal-Ligand Polyhedra
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Huddy TF, Hsia Y, Kibler RD, Xu J, Bethel N, Nagarajan D, Redler R, Leung PJY, Weidle C, Courbet A, Yang EC, Bera AK, Coudray N, Calise SJ, Davila-Hernandez FA, Han HL, Carr KD, Li Z, McHugh R, Reggiano G, Kang A, Sankaran B, Dickinson MS, Coventry B, Brunette TJ, Liu Y, Dauparas J, Borst AJ, Ekiert D, Kollman JM, Bhabha G, Baker D. Blueprinting extendable nanomaterials with standardized protein blocks. Nature. 2024 Mar;627(8005):898-904. PMID:38480887 doi:10.1038/s41586-024-07188-4
- ↑ Brunette et al. (David Baker lab), [https://doi.org/10.1101/2025.01.29.635581 A Multivalent Pan-Ebolavirus Nanoparticle Vaccine Provides Protection in Rodents from Lethal Infection by Adapted Zaire and Sudan Viruses], bioRXiv Preprint, February, 2005.
- ↑ An amino-terminal histidine tag was not resolved in the electron density map of 8g9j, and thus is missing in the structure depicted.
- ↑ de Haas RJ, Brunette N, Goodson A, Dauparas J, Yi SY, Yang EC, Dowling Q, Nguyen H, Kang A, Bera AK, Sankaran B, de Vries R, Baker D, King NP. Rapid and automated design of two-component protein nanomaterials using ProteinMPNN. Proc Natl Acad Sci U S A. 2024 Mar 26;121(13):e2314646121. PMID:38502697 doi:10.1073/pnas.2314646121
- ↑ Sumida KH, Núñez-Franco R, Kalvet I, Pellock SJ, Wicky BIM, Milles LF, Dauparas J, Wang J, Kipnis Y, Jameson N, Kang A, De La Cruz J, Sankaran B, Bera AK, Jiménez-Osés G, Baker D. Improving Protein Expression, Stability, and Function with ProteinMPNN. J Am Chem Soc. 2024 Jan 9. PMID:38194293 doi:10.1021/jacs.3c10941
- ↑ Dauparas J, Anishchenko I, Bennett N, Bai H, Ragotte RJ, Milles LF, Wicky BIM, Courbet A, de Haas RJ, Bethel N, Leung PJY, Huddy TF, Pellock S, Tischer D, Chan F, Koepnick B, Nguyen H, Kang A, Sankaran B, Bera AK, King NP, Baker D. Robust deep learning-based protein sequence design using ProteinMPNN. Science. 2022 Sep 15:eadd2187. doi: 10.1126/science.add2187. PMID:36108050 doi:http://dx.doi.org/10.1126/science.add2187
- ↑ The five modules shown are from the following assemblies that can be downloaded as PDB files from supplementary materials of Huddy et al,, 2024: 90° from strut_C6_16. Branch from TT_rail+_tie+. Curve from R20A. Handshake 90° from cage_O4_32. Handshake obtuse angle from cage_I3_8.
- ↑ The assemblies shown can be downloaded as PDB files from supplementary materials of Huddy et al,, 2024.
- ↑ Wang S, Favor A, Kibler R, Lubner J, Borst AJ, Coudray N, Redler RL, Chiang HT, Sheffler W, Hsia Y, Li Z, Ekiert DC, Bhabha G, Pozzo LD, Baker D. Bond-centric modular design of protein assemblies. bioRxiv [Preprint]. 2024 Oct 12:2024.10.11.617872. PMID:39416012 doi:10.1101/2024.10.11.617872