We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1847
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
===What are Minibinders?=== | ===What are Minibinders?=== | ||
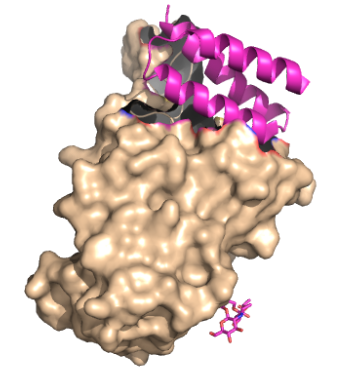
| - | Minibinders are small proteins that bind to the spike protein that is involved in the viral infection pathway for SARS-CoV-2. | + | Minibinders are small proteins that bind to the spike protein that is involved in the viral infection pathway for SARS-CoV-2. These mini proteins target the interaction between SARS-CoV-2 spike protein and ACE2 receptor as an effective therapeutic strategy <ref name="Longxing">PMID:34192518</ref>. The demand for SARS-CoV-2 therapeutics is high, and the promise these minibinders have shown is great. |
===COVID-19 Viral Infection Interruption=== | ===COVID-19 Viral Infection Interruption=== | ||
==Subheading 2== | ==Subheading 2== | ||
Revision as of 18:49, 27 March 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Contents |
Structure
Introduction
What are Minibinders?
Minibinders are small proteins that bind to the spike protein that is involved in the viral infection pathway for SARS-CoV-2. These mini proteins target the interaction between SARS-CoV-2 spike protein and ACE2 receptor as an effective therapeutic strategy [1]. The demand for SARS-CoV-2 therapeutics is high, and the promise these minibinders have shown is great.
COVID-19 Viral Infection Interruption
Subheading 2
| |||||||||||
References
- ↑ 1.0 1.1 Case JB, Chen RE, Cao L, Ying B, Winkler ES, Johnson M, Goreshnik I, Pham MN, Shrihari S, Kafai NM, Bailey AL, Xie X, Shi PY, Ravichandran R, Carter L, Stewart L, Baker D, Diamond MS. Ultrapotent miniproteins targeting the SARS-CoV-2 receptor-binding domain protect against infection and disease. Cell Host Microbe. 2021 Jul 14;29(7):1151-1161.e5. PMID:34192518 doi:10.1016/j.chom.2021.06.008
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644