Sandbox Reserved 1847
From Proteopedia
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
===What are Minibinders?=== | ===What are Minibinders?=== | ||
| - | + | These mini proteins target the interaction between ACE2 and COVID-19 spike protein <ref name="Longxing">PMID:32907861</ref>. The mini binders are small proteins carefully designed to bind to the COVID-19 spike protein with a greater affinity than ACE2 <ref name="Longxing">PMID:32907861</ref>. These mini binders were able to reduce the viral burden of SARS-CoV-2 in mice <ref name="Case">PMID:34192518</ref>. These proteins were de novo (from scratch) designs to mimic the ACE2 helix, but have a lower dissociation constant, yielding a greater affinity for the spike protein <ref name="Longxing">PMID:32907861</ref>. | |
===COVID-19 Disease Pathway=== | ===COVID-19 Disease Pathway=== | ||
| - | Understanding the pathway | + | Understanding the pathway of the COVID-19 virus is essential to understanding the mechanism in which the virus’ surface proteins attach to the mini binders. The COVID-19 virus has spike proteins on its surface that bind to the host cell receptor known as ACE2 <ref name="Beyerstedt">PMID:33389262</ref>. When the spike protein binds to ACE2, this allows the virus to anchor to the host cell, permitting the endocytosis of the virus, allowing it to translate viral proteins, which will ultimately result in an immune response <ref name="Beyerstedt">PMID:33389262</ref>. The ACE2 protein regulates the Renin-angiotensin-aldosterone system, which an imbalance, due to the spike protein, can lead to the negative clinical effects that are often observed in patients with COVID-19, but especially those with hypertension, diabetes and cardiovascular diseases <ref name="Beyerstedt">PMID:33389262</ref>. |
===COVID-19 Viral Infection Interruption=== | ===COVID-19 Viral Infection Interruption=== | ||
| - | The | + | The primary goal of the mini binders is to prevent the , through binding to the COVID-19 spike protein and the mini binders are incredibly effective at doing such <ref name="Case">PMID:34192518</ref>. Because the mini binders have a greater binding affinity than ACE2 for the spike protein, they are able to effectively prevent the entry of the virus and ultimately prevent an immune response <ref name="Longxing">PMID:32907861</ref>. |
| + | |||
==Design== | ==Design== | ||
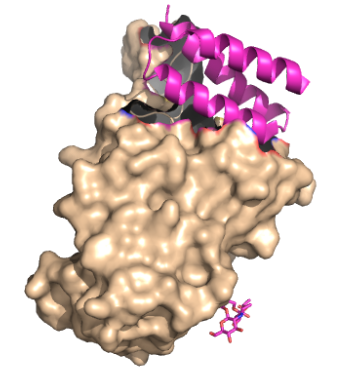
| - | These | + | These mini binders, LCB1 and AHB2, were designed from “scratch” (de novo) with the intention to mimic the binding of ACE2 to spike protein (insert citation). Using Rotamer Interaction Field (RIF) docking, the proteins were able to make the most efficient bonding using the ACE2 spike protein binding interface (insert citation). Using Site Saturation Mutagenesis (SSM), every residue in the minibinder’s helix scaffold will be substituted with each of the 20 amino acids, one at a time (insert citation). Forming SSM libraries, each of the libraries converged on a small number of closely related sequences, and from these libraries, the design was selected for LCB1 and AHB2 to find the sequence that yields a protein with a high affinity for the spike proteins receptor binding domain (insert). |
<StructureSection load='7JZU' size='340' side='right' caption='Caption for this structure' scene='10/1075249/Lcb1_spikerbd/1'> | <StructureSection load='7JZU' size='340' side='right' caption='Caption for this structure' scene='10/1075249/Lcb1_spikerbd/1'> | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
Revision as of 19:40, 3 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Contents |
Structure
Introduction
What are Minibinders?
These mini proteins target the interaction between ACE2 and COVID-19 spike protein [1]. The mini binders are small proteins carefully designed to bind to the COVID-19 spike protein with a greater affinity than ACE2 [1]. These mini binders were able to reduce the viral burden of SARS-CoV-2 in mice [2]. These proteins were de novo (from scratch) designs to mimic the ACE2 helix, but have a lower dissociation constant, yielding a greater affinity for the spike protein [1].
COVID-19 Disease Pathway
Understanding the pathway of the COVID-19 virus is essential to understanding the mechanism in which the virus’ surface proteins attach to the mini binders. The COVID-19 virus has spike proteins on its surface that bind to the host cell receptor known as ACE2 [3]. When the spike protein binds to ACE2, this allows the virus to anchor to the host cell, permitting the endocytosis of the virus, allowing it to translate viral proteins, which will ultimately result in an immune response [3]. The ACE2 protein regulates the Renin-angiotensin-aldosterone system, which an imbalance, due to the spike protein, can lead to the negative clinical effects that are often observed in patients with COVID-19, but especially those with hypertension, diabetes and cardiovascular diseases [3].
COVID-19 Viral Infection Interruption
The primary goal of the mini binders is to prevent the , through binding to the COVID-19 spike protein and the mini binders are incredibly effective at doing such [2]. Because the mini binders have a greater binding affinity than ACE2 for the spike protein, they are able to effectively prevent the entry of the virus and ultimately prevent an immune response [1].
Design
These mini binders, LCB1 and AHB2, were designed from “scratch” (de novo) with the intention to mimic the binding of ACE2 to spike protein (insert citation). Using Rotamer Interaction Field (RIF) docking, the proteins were able to make the most efficient bonding using the ACE2 spike protein binding interface (insert citation). Using Site Saturation Mutagenesis (SSM), every residue in the minibinder’s helix scaffold will be substituted with each of the 20 amino acids, one at a time (insert citation). Forming SSM libraries, each of the libraries converged on a small number of closely related sequences, and from these libraries, the design was selected for LCB1 and AHB2 to find the sequence that yields a protein with a high affinity for the spike proteins receptor binding domain (insert).
| |||||||||||
|