This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Morphs
From Proteopedia
(→Why Morph? - polishing) |
(→Why Morph? - polishing) |
||
| Line 3: | Line 3: | ||
==Why Morph?== | ==Why Morph?== | ||
| - | + | <table align='right' border='1' width='285' cellpadding='10' bgcolor='#d0d0d0' hspace='8'><tr><td> | |
| + | [[Image:Mage_hb.gif]]</td></tr><tr><td>Toggling between the carbonmonoxy and deoxy conformations of heme in hemoglobin. Convergent stereo snapshot from a Kinemage. (Stops after 25 cycles; reload this page to restart the toggling.)</td></tr></table> | ||
The purpose of molecular morphing is to smooth the visual transition between two molecular conformations, making it easier to see and understand the structural differences between them. | The purpose of molecular morphing is to smooth the visual transition between two molecular conformations, making it easier to see and understand the structural differences between them. | ||
In contrast, predicting the actual trajectory through which a conformational change occurs is rarely, if ever, the goal of a morph. | In contrast, predicting the actual trajectory through which a conformational change occurs is rarely, if ever, the goal of a morph. | ||
Some proteins perform their functions without major conformational changes. On the other hand, some proteins must undergo major changes in secondary, tertiary, or quaternary structure in order to perform their functions. In quite a few cases, investigators have succeeded in obtaining empirically determined structures for a protein in two or more conformations. The challenge for visualization is then to be able to follow the changes in each region between the two conformations. | Some proteins perform their functions without major conformational changes. On the other hand, some proteins must undergo major changes in secondary, tertiary, or quaternary structure in order to perform their functions. In quite a few cases, investigators have succeeded in obtaining empirically determined structures for a protein in two or more conformations. The challenge for visualization is then to be able to follow the changes in each region between the two conformations. | ||
| - | <table align='right' border='1' width='285' cellpadding='10' bgcolor='#d0d0d0'><tr><td> | ||
| - | [[Image:Mage_hb.gif]]</td></tr><tr><td>Toggling between the carbonmonoxy and deoxy conformations of heme in hemoglobin. Convergent stereo snapshot from a Kinemage. (Stops after 25 cycles; reload this page to restart the toggling.)</td></tr></table> | ||
When the differences are small, simply toggling an image between the two states is adequate. [http://kinemage.biochem.duke.edu/ David Richardson's MAGE], first available in 1992<ref>[http://history.molviz.org History of Macromolecular Visualization]</ref> supports visual toggling between macromolecular conformations. Hundreds of interactive molecular structure tutorials called kinemages take advantage of this capability. At the right are snapshots of hemoglobin toggled in MAGE. MAGE is available in java applet form, and is an option for displaying molecular scenes in Proteopedia<ref>For sample pages that use the MAGE applet in Proteopedia, see [[Hemoglobin]] and [[Ribulose-1,5-bisphosphate carboxylase/oxygenase]]</ref>. | When the differences are small, simply toggling an image between the two states is adequate. [http://kinemage.biochem.duke.edu/ David Richardson's MAGE], first available in 1992<ref>[http://history.molviz.org History of Macromolecular Visualization]</ref> supports visual toggling between macromolecular conformations. Hundreds of interactive molecular structure tutorials called kinemages take advantage of this capability. At the right are snapshots of hemoglobin toggled in MAGE. MAGE is available in java applet form, and is an option for displaying molecular scenes in Proteopedia<ref>For sample pages that use the MAGE applet in Proteopedia, see [[Hemoglobin]] and [[Ribulose-1,5-bisphosphate carboxylase/oxygenase]]</ref>. | ||
Revision as of 00:47, 5 October 2008
This page is in a rough and incomplete state. I plan to get it into acceptable shape soon. Eric Martz 04:35, 15 June 2008 (IDT)
Contents |
Why Morph?
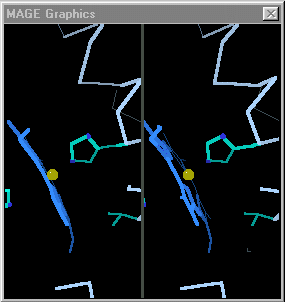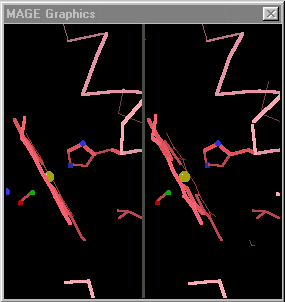 |
| Toggling between the carbonmonoxy and deoxy conformations of heme in hemoglobin. Convergent stereo snapshot from a Kinemage. (Stops after 25 cycles; reload this page to restart the toggling.) |
The purpose of molecular morphing is to smooth the visual transition between two molecular conformations, making it easier to see and understand the structural differences between them. In contrast, predicting the actual trajectory through which a conformational change occurs is rarely, if ever, the goal of a morph.
Some proteins perform their functions without major conformational changes. On the other hand, some proteins must undergo major changes in secondary, tertiary, or quaternary structure in order to perform their functions. In quite a few cases, investigators have succeeded in obtaining empirically determined structures for a protein in two or more conformations. The challenge for visualization is then to be able to follow the changes in each region between the two conformations. When the differences are small, simply toggling an image between the two states is adequate. David Richardson's MAGE, first available in 1992[1] supports visual toggling between macromolecular conformations. Hundreds of interactive molecular structure tutorials called kinemages take advantage of this capability. At the right are snapshots of hemoglobin toggled in MAGE. MAGE is available in java applet form, and is an option for displaying molecular scenes in Proteopedia[2].
On the other hand, when the conformational changes are large, toggling between two very different conformations leaves the eye unable to grasp what has happened. This is when morphing is useful for seeing and understanding the differences between conformations.
Examples of Morphs
Morphs in Proteopedia
- Avian Influenza Neuraminidase, Tamiflu and Relenza shows a morph of the induced fit of N1 to Tamiflu.
- Proton Channels includes a morph of a transmembrane channel opening and closing.
- Recoverin, a calcium-activated myristoyl switch includes a morph of the calcium-activated conformational change that expels the myristate from within a protein domain, converting the protein from soluble to membrane anchored with consequent redistribution of its activity.
Morphs Elsewhere
Visualizing Morphs
(to be added)
Morphing Methods
Linear Interpolation
In this method, a series of intermediate models are created in which each atom moves in a straight line from is initial position to its final position. This type of morphing is relatively easy to perform, and often suffices, but also has a number of limitations. When animated, the interpolated intermediate conformations often greatly help in visualizing the differences between the initial and final empirical models. However, otherwise they are otherwise meaningless. Bond lengths and angles become unrealistic, domains may artifactually shrink, expand, or distort, and chains may even pass through each other during the interpolated movements. When there are substantial changes in secondary structure or large movements of domains, linear interpolation becomes unsatisfactory.
A toggler is implemented here for Recoverin. Despite such features as the ability to highlight any arbitrary range of residues, it remains too difficult to follow the changes in secondary and tertiary structure during the large jumps in position. Consequently, a series of intermediate conformations were generated by linear interpolation of alpha carbon positions. This backbone trace "morph" makes it much easier to follow the relations between the two empirical conformations. However, Linear interpolation morphing was first employed by Vonrhein, Schlauderer & Schultz (1995) to create movies of the results of substrate binding to nucleoside monophosphate kinases.
Chemically Possible Intermediates
Plausible intermediate conformations have been calculated by Mark Gerstein and Werner Krebs at Yale University, using a combination of linear interpolation and molecular dynamics. Gerstein has generously made his interpolation data available, and in some cases they are here displayed in this Chime-based Protein Morpher interface. At the Gerstein Lab Homepage you will find movies of plausible morphs in animated GIF's, Quicktime, and VRML. Another source of "chemically reasonable" morphs is the Biomolecular Morphing site by Gerard J. Kleywegt at Uppsala University, Sweden.
References, LInks, and Notes
- ↑ History of Macromolecular Visualization
- ↑ For sample pages that use the MAGE applet in Proteopedia, see Hemoglobin and Ribulose-1,5-bisphosphate carboxylase/oxygenase
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Wayne Decatur, Karsten Theis, Joel L. Sussman, Angel Herraez, David Canner, Eran Hodis
