User:Nathan Roy
From Proteopedia
| Line 76: | Line 76: | ||
| + | <applet load='1A1T' size='400' frame='true' align='left' caption='Figure 5. NC in complex with Viral SL3 element' /> | ||
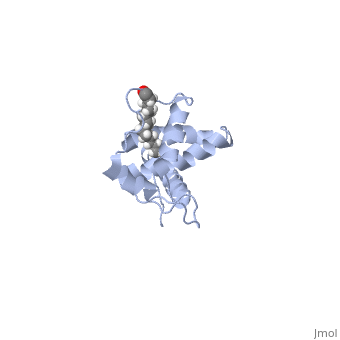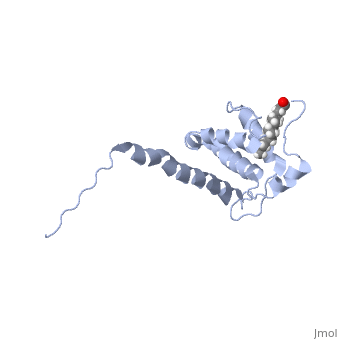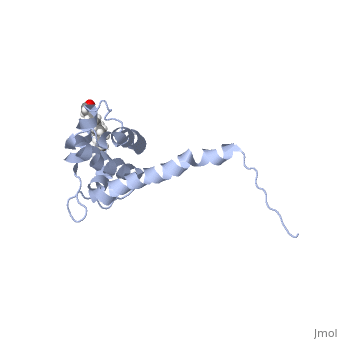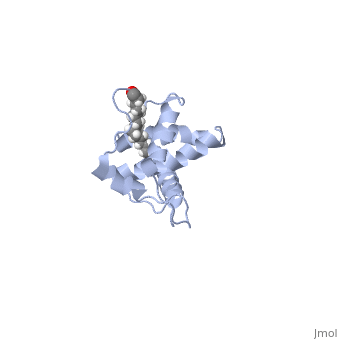
| - | + | As we have seen, Gag is responsible for correct targeting of viral assembly to discreet sites on the plasma membrane, and viral capsid structure assembly into organized virus particles. However, Gag is also responsible for packaging of the viral RNA into budding virions, and this function in executed by the NC domain. The 5' LTR of HIV-1 genomic RNA contains a recognition element called the psi element. All retroviruses contain some type of psi element in order to get specific packaging of viral genomic RNA within the budding particles, and in the case of HIV-1, the psi element is 120 bases, and contains 4 stem-loop structures. Although the psi element can be quite variable, the SL3 loop is highly conserved within HIV-1 strains. Figure 5 shows the NC domain of HIV-1 NL4-3 in complex with the SL3 loop of the viral psi element. | |
| - | + | ||
Revision as of 19:44, 22 April 2009
User:Nathan Roy/Sandbox 1I am a graduate student at the University of Vermont, studying the dynamics of HIV-1 cell to cell transmission and HIV-1 induced syncytia formation. Our wonderful professor, Dr. Steven Everse, has commissioned us (his students from his BioChem 351 course) to create a page describing the structure of a protein that interests us. I have chosen the HIV-1 gag protein, and more specifically, the MA and CA domains.
HIV-1 Gag
The HIV-1 Gag protein is the major structural protein required for virus assembly. It is synthesized as a polyprotein in the cytosol of an infected cell, and contains four functional segments; MA, CA (NTD and CTD), NC, and p6. The NC region is flanked by two "spacer" segments, denoted SP1 and SP2. The polyprotein is all alpha helical, except the NC region, which is composed of two RNA interacting zinc knuckle domains. Gag is often referred to as an "assembly machine", because expression of Gag alone is sufficient to produce budding virus-like particles (VLP's), due to multimerization of roughly 2000 Gag molecules per virion. Here, we will take a closer look at the MA, CA, and NC domains, and how the structural components of these domains aid in the assembly of virus particles. Viral particles can be classified as immature (pre-budding and non-infectious), and mature (post-budding and infectious), and this exchange is mediated by the HIV-1 protease(LINK). Upon viral budding, Gag is cleaved by the HIV-1 protease at multiple sites, thus possibly changing many of the structural interactions that make up the "immature" particle. For simplicity, we will only be discussing the immature formation of Gag on the plasma membrane of infected cells, as it coordinates organized viral budding. Also, it is thought that Gag forms a hexamer structure upon virus assembly, but because of the difficulties encountered by attempting to crystallize a multimeric structure, the exact formation of the hexamer is still up for debate.
Matrix (MA)
|
|
Capsid (CA)
|
|
Nucleocapsid (NC)
|
As we have seen, Gag is responsible for correct targeting of viral assembly to discreet sites on the plasma membrane, and viral capsid structure assembly into organized virus particles. However, Gag is also responsible for packaging of the viral RNA into budding virions, and this function in executed by the NC domain. The 5' LTR of HIV-1 genomic RNA contains a recognition element called the psi element. All retroviruses contain some type of psi element in order to get specific packaging of viral genomic RNA within the budding particles, and in the case of HIV-1, the psi element is 120 bases, and contains 4 stem-loop structures. Although the psi element can be quite variable, the SL3 loop is highly conserved within HIV-1 strains. Figure 5 shows the NC domain of HIV-1 NL4-3 in complex with the SL3 loop of the viral psi element.