User:Karen Lee/Sandbox 1
From Proteopedia
m |
|||
| Line 1: | Line 1: | ||
==Replication Termination== | ==Replication Termination== | ||
| - | Replication is an essential process in all cells. The process copies the chromosomal DNA of the organism to provide the extra copy needed in cell division and is therefore critical in the biological inheritance of genes. In cells with circular chromosomes, replication starts from a single origin proceed with two replication forks moving in opposite directions. | + | Replication is an essential process in all cells. The process copies the chromosomal DNA of the organism to provide the extra copy needed in cell division and is therefore critical in the biological inheritance of genes. In cells with circular chromosomes, replication starts from a single origin proceed with two replication forks moving in opposite directions. It should follow that this process must be terminated or multiple copies of the chromosome would be made. |
| - | The process of terminating replication is performed by replication termination proteins. These proteins bind to specific sequences in the DNA, called ''Ter'' sites. This binding provides a physical blockage in the DNA that stops the replication machinery. | + | The process of terminating replication is performed by replication termination proteins. These proteins bind to specific sequences in the DNA, called ''Ter'' sites. This binding provides a physical blockage in the DNA that stops the replication machinery. In ''B. subtilis'', the termination protein is called Replication Terminator Protein (RTP), and in ''E. coli'' it is Termination Utilisation Substance (Tus). |
| - | In each circular chromosome, there are two sets of ''Ter'' sites that appear roughly opposite to the origin of replication. One set blocks the clockwise replication fork while the other traps the anti-clockwise replication fork. | + | In each circular chromosome, there are two sets of ''Ter'' sites that appear roughly opposite to the origin of replication. One set blocks the clockwise replication fork while the other traps the anti-clockwise replication fork. Interestingly, both these proteins bind to DNA in such a way that they terminate the replication fork in one direction, but simultaneously allow the replication fork to continue in the other direction. |
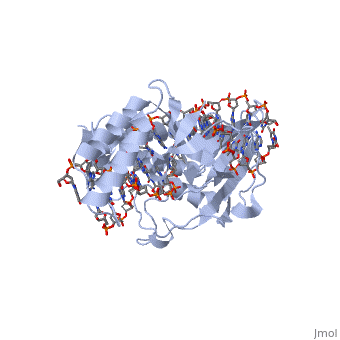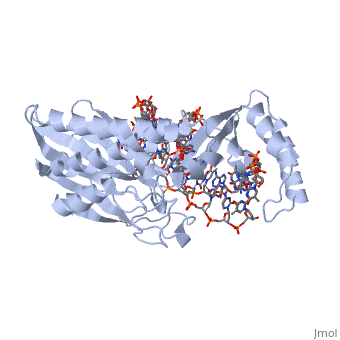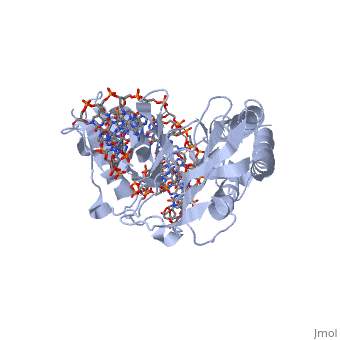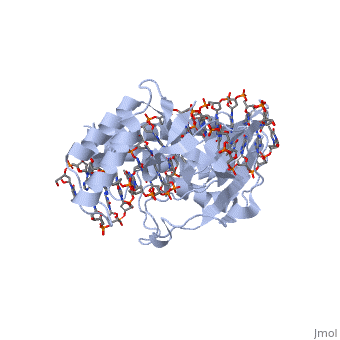
| - | + | ==RTP== | |
| + | <Structure load='1f4k' size='400' frame='true' align='right' caption='An RTP dimer bound to the B-site Ter sequence' scene='Insert optional scene name here' /> | ||
| - | + | ===Structure of RTP=== | |
| + | A single RTP monomer consists of four <scene name='User:Karen_Lee/Sandbox_1/Rtp_alpha_helices/3'>α-helices</scene>, three <scene name='User:Karen_Lee/Sandbox_1/Rtp_beta_sheets/2'>β-strands</scene> and an unstructured <scene name='User:Karen_Lee/Sandbox_1/Rtp_n_terminal/1'>N-terminal</scene> domain. The <scene name='User:Karen_Lee/Sandbox_1/Rtp_alpha_helix_3_dna/2'>α3 helix</scene> binds to DNA by <scene name='User:Karen_Lee/Sandbox_1/Rtp_alpha_helix_3_dna/1'>inserting</scene> into the major groove of Ter sites, while the <scene name='User:Karen_Lee/Sandbox_1/Rtp_beta_strand_dna/1'>β2-strand</scene> interacts with the minor groove. The N-terminal arm also binds to the Ter site<ref>Pai, S. K., Bussiere, D. E., Wang, F., Hutchinson, C. A., White, S. W. & Bastia, D. (1996) The structure and function of the replication terminator protein of Bacillus subtilis: identification of the ‘winged helix’ DNA-binding domain. ''EMBO J.'' '''15(12)''', 3164-3173.</ref>. | ||
| - | + | RTP conforms to the classic winged-helix motif, a compact α/β structure with αβααββ topology where "<scene name='User:Karen_Lee/Sandbox_1/Wing/1'>wings</scene>" project from the loop between β2 and β3-strands<ref>Gajiwala, K. S. & Burley, S. K. (2000) Winged Helix proteins. ''Curr. Opin. Struct. Biol.'' '''10''', 110-116.</ref>. However, each monomer lacks a well-formed β1-strand (named β1-loop) and has an additional α4-helix following the β3-strand<ref name="Vivian">Vivian, J. P., Porter, C. J., Wilce, J. A. & Wilce, M. C. J. (2007) An Asymmetric Structure of the Bacillus subtilis Replication Terminator Protein in Complex with DNA. ''J. Mol. Biol.'' '''370''', 481-491.</ref>. | |
| - | < | + | |
| - | + | (<scene name='User:Karen_Lee/Sandbox_1/Rtp_revert/1'>Click to revert back to original view</scene>.) | |
| - | + | ||
| - | <scene name='User:Karen_Lee/Sandbox_1/ | + | ===Key Features of RTP=== |
| + | |||
| + | Two different RTP B sites have been found to interact with RTP. These are the symmetrical RTP B (sRB) site from TerI (the first Ter site that the clockwise replication fork encounters) and the native RTP B (nRB) site from TerI. These sequences differ only in 6 base pairs – three at the downstream end and three at the upstream end. The downstream changes have no bearing on the structure of RTP since the protein binds the downstream region in both sRB and nRB sequences with similar conformation. Additionally, no base-specific interactions are made in this region. However, the three upstream changes are all located in the major groove of the dsDNA. This is where the α3 helix binds which underlies RTP binding specificity<ref name="Vivian" />. Therefore, in binding of RTP monomers to the Ter site, there is a differential binding affinity in A and B sites, due to changes in the nRB site. | ||
| + | |||
| + | This leads into the fact that RTP binds asymmetrically across the nRB site and therefore allows the complex to act as a polar barrier to the replication fork. When the fork approaches the B site (with tight RTP-DNA binding) the fork is unable to progress and is paused. Approaching from the A site does not impede its progress. This polarity can be explained by both the differential binding affinity as explained above and the cooperative binding affect of the RTP monomers. The complex formed between an RTP molecule and the B site facilitates cooperative binding of another RTP monomer to the A site to form a complete RTP-Ter complex<ref name="Vivian" />. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Tus== | ||
| + | <Structure load='1ecr' size='400' frame='true' align='left' caption='TUS complexed with DNA' /> | ||
| + | |||
| + | ===Structure of Tus=== | ||
| + | The Tus protein works as a monomer, each monomer consists of two domains, the amino and carboxy domains. The overall structure of Tus is made up of two <scene name='User:Karen_Lee/Sandbox_1/Tus_withdna_alphahelices/1'>two α-helical regions</scene> separated by central twisted anti-parallel <scene name='User:Karen_Lee/Sandbox_1/Tus_withdna_betasheets/1'>β-strands</scene>. These structures together form a <scene name='User:Karen_Lee/Sandbox_1/Tus_nodna/1'>positively charged central cleft</scene> that accommodates a B-form Ter <scene name='User:Karen_Lee/Sandbox_1/Tus_withdna/1'>DNA duplex</scene>. This binding involves two β-strands involes contacting the bases and sugar-phosphate back bones from the major groove of the Ter DNA. Upon Tus binding, the major groove becomes deeper and the minor groove is significantly deeper, causing the DNA to be slightly bent. | ||
| - | ==Key Features of RTP== | ||
| - | Two different RTP B sites have been found to interact with RTP. These are the symmetrical RTP B (sRB) site from TerI (the first Ter site that the clockwise replication fork encounters) and the native RTP B (nRB) site from TerI. These sequences differ only in 6 base pairs – three at the downstream end and three at the upstream end. The downstream changes have no bearing on the structure of RTP since the protein binds the downstream region in both sRB and nRB sequences with similar conformation. Additionally, no base-specific interactions are made in this region. However, the three upstream changes are all located in the major groove of the dsDNA. This is where the α3 helix binds which underlies RTP binding specificity<ref>Vivian, J. P., Porter, C. J., Wilce, J. A. & Wilce, M. C. J. (2007) An Asymmetric Structure of the Bacillus subtilis Replication Terminator Protein in Complex with DNA. J. Mol. Biol. 370, 481-491.</ref>. Therefore, in binding of RTP monomers to the Ter site, there is a differential binding affinity in A and B sites, due to changes in the nRB site. | ||
| - | This leads into the fact that RTP binds asymmetrically across the nRB site and therefore allows the complex to act as a polar barrier to the replication fork. When the fork approaches the B site (with tight RTP-DNA binding) the fork is unable to progress and is paused. Approaching from the A site does not impede its progress. This polarity can be explained by both the differential binding affinity as explained above and the cooperative binding affect of the RTP monomers. The complex formed between an RTP molecule and the B site facilitates cooperative binding of another RTP monomer to the A site to form a complete RTP-Ter complex<ref>Vivian, J. P., Porter, C. J., Wilce, J. A. & Wilce, M. C. J. (2007) An Asymmetric Structure of the Bacillus subtilis Replication Terminator Protein in Complex with DNA. J. Mol. Biol. 370, 481-491.</ref>. | ||
| Line 30: | Line 42: | ||
| - | =TUS= | ||
| - | <Structure load='1ecr' size='400' frame='true' align='left' caption='TUS complexed with DNA' scene='User:Karen_Lee/Sandbox_1/Freeze_frame/1' /> | ||
| - | == Structure of Tus == | ||
| - | The Tus protein works as a monomer, each monomer consists of two domains, the amino and carboxy domains. The overall structure of Tus is made up of two <scene name='User:Karen_Lee/Sandbox_1/Tus_withdna_alphahelices/1'>two α-helical regions</scene> separated by central twisted anti-parallel <scene name='User:Karen_Lee/Sandbox_1/Tus_withdna_betasheets/1'>β-strands</scene>. These structures together form a <scene name='User:Karen_Lee/Sandbox_1/Tus_nodna/1'>positively charged central cleft</scene> that accommodates a B-form Ter <scene name='User:Karen_Lee/Sandbox_1/Tus_withdna/1'>DNA duplex</scene>. This binding involves two β-strands involes contacting the bases and sugar-phosphate back bones from the major groove of the Ter DNA. Upon Tus binding, the major groove becomes deeper and the minor groove is significantly deeper, causing the DNA to be slightly bent. | ||
Revision as of 06:09, 14 May 2011
Contents |
Replication Termination
Replication is an essential process in all cells. The process copies the chromosomal DNA of the organism to provide the extra copy needed in cell division and is therefore critical in the biological inheritance of genes. In cells with circular chromosomes, replication starts from a single origin proceed with two replication forks moving in opposite directions. It should follow that this process must be terminated or multiple copies of the chromosome would be made.
The process of terminating replication is performed by replication termination proteins. These proteins bind to specific sequences in the DNA, called Ter sites. This binding provides a physical blockage in the DNA that stops the replication machinery. In B. subtilis, the termination protein is called Replication Terminator Protein (RTP), and in E. coli it is Termination Utilisation Substance (Tus).
In each circular chromosome, there are two sets of Ter sites that appear roughly opposite to the origin of replication. One set blocks the clockwise replication fork while the other traps the anti-clockwise replication fork. Interestingly, both these proteins bind to DNA in such a way that they terminate the replication fork in one direction, but simultaneously allow the replication fork to continue in the other direction.
RTP
|
Structure of RTP
A single RTP monomer consists of four , three and an unstructured domain. The binds to DNA by into the major groove of Ter sites, while the interacts with the minor groove. The N-terminal arm also binds to the Ter site[1].
RTP conforms to the classic winged-helix motif, a compact α/β structure with αβααββ topology where "" project from the loop between β2 and β3-strands[2]. However, each monomer lacks a well-formed β1-strand (named β1-loop) and has an additional α4-helix following the β3-strand[3].
(.)
Key Features of RTP
Two different RTP B sites have been found to interact with RTP. These are the symmetrical RTP B (sRB) site from TerI (the first Ter site that the clockwise replication fork encounters) and the native RTP B (nRB) site from TerI. These sequences differ only in 6 base pairs – three at the downstream end and three at the upstream end. The downstream changes have no bearing on the structure of RTP since the protein binds the downstream region in both sRB and nRB sequences with similar conformation. Additionally, no base-specific interactions are made in this region. However, the three upstream changes are all located in the major groove of the dsDNA. This is where the α3 helix binds which underlies RTP binding specificity[3]. Therefore, in binding of RTP monomers to the Ter site, there is a differential binding affinity in A and B sites, due to changes in the nRB site.
This leads into the fact that RTP binds asymmetrically across the nRB site and therefore allows the complex to act as a polar barrier to the replication fork. When the fork approaches the B site (with tight RTP-DNA binding) the fork is unable to progress and is paused. Approaching from the A site does not impede its progress. This polarity can be explained by both the differential binding affinity as explained above and the cooperative binding affect of the RTP monomers. The complex formed between an RTP molecule and the B site facilitates cooperative binding of another RTP monomer to the A site to form a complete RTP-Ter complex[3].
Tus
|
Structure of Tus
The Tus protein works as a monomer, each monomer consists of two domains, the amino and carboxy domains. The overall structure of Tus is made up of two separated by central twisted anti-parallel . These structures together form a that accommodates a B-form Ter . This binding involves two β-strands involes contacting the bases and sugar-phosphate back bones from the major groove of the Ter DNA. Upon Tus binding, the major groove becomes deeper and the minor groove is significantly deeper, causing the DNA to be slightly bent.
References
- ↑ Pai, S. K., Bussiere, D. E., Wang, F., Hutchinson, C. A., White, S. W. & Bastia, D. (1996) The structure and function of the replication terminator protein of Bacillus subtilis: identification of the ‘winged helix’ DNA-binding domain. EMBO J. 15(12), 3164-3173.
- ↑ Gajiwala, K. S. & Burley, S. K. (2000) Winged Helix proteins. Curr. Opin. Struct. Biol. 10, 110-116.
- ↑ 3.0 3.1 3.2 Vivian, J. P., Porter, C. J., Wilce, J. A. & Wilce, M. C. J. (2007) An Asymmetric Structure of the Bacillus subtilis Replication Terminator Protein in Complex with DNA. J. Mol. Biol. 370, 481-491.