This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Journal:JBIC:13
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
The high resolution crystal structure of CODH-II from ''Carboxydothermus hydrogenoformans'' (d<sub>min</sub> = 1.28 Å) revealed <scene name='Journal:JBIC:13/Cv/16'>novel type of the Ni-carbon bond at the C-cluster upon binding of n-butylisocyanate</scene> (the product of n-BIC oxidation), in which the <scene name='Journal:JBIC:13/Cv/17'>Ni complex with the n-butylisocyanate ligand shows a distorted tetrahedral geometry</scene> rather than the <scene name='Journal:JBIC:13/Al/2'>square planar geometry observed in the CO2-bound</scene> ([[3b52]]) and <scene name='Journal:JBIC:13/Al/3'>CN-bound</scene> states ([[3i39]]). <scene name='Journal:JBIC:13/Cv1/1'>The hydroxyl group of n-butylisocyanate bound to the Ni atom is in a proper distance and orientation to form hydrogen bonds with His93 and the H2O/OH- ligand on Fe1A</scene>, while <scene name='Journal:JBIC:13/Cv1/2'>apolar amino acid side chains, Ile567 and Ala564, accommodate the long chain alkyl group of n-butylisocyanate</scene>. A superposition of the CODH-II structures with different ligands bound to the C-cluster reveals a flexible coordination and geometry for the Ni-Fe1 dyad, while the [Fe<sub>3</sub>S<sub>4</sub>] moiety of C-cluster remains unchanged in position. | The high resolution crystal structure of CODH-II from ''Carboxydothermus hydrogenoformans'' (d<sub>min</sub> = 1.28 Å) revealed <scene name='Journal:JBIC:13/Cv/16'>novel type of the Ni-carbon bond at the C-cluster upon binding of n-butylisocyanate</scene> (the product of n-BIC oxidation), in which the <scene name='Journal:JBIC:13/Cv/17'>Ni complex with the n-butylisocyanate ligand shows a distorted tetrahedral geometry</scene> rather than the <scene name='Journal:JBIC:13/Al/2'>square planar geometry observed in the CO2-bound</scene> ([[3b52]]) and <scene name='Journal:JBIC:13/Al/3'>CN-bound</scene> states ([[3i39]]). <scene name='Journal:JBIC:13/Cv1/1'>The hydroxyl group of n-butylisocyanate bound to the Ni atom is in a proper distance and orientation to form hydrogen bonds with His93 and the H2O/OH- ligand on Fe1A</scene>, while <scene name='Journal:JBIC:13/Cv1/2'>apolar amino acid side chains, Ile567 and Ala564, accommodate the long chain alkyl group of n-butylisocyanate</scene>. A superposition of the CODH-II structures with different ligands bound to the C-cluster reveals a flexible coordination and geometry for the Ni-Fe1 dyad, while the [Fe<sub>3</sub>S<sub>4</sub>] moiety of C-cluster remains unchanged in position. | ||
| - | + | ||
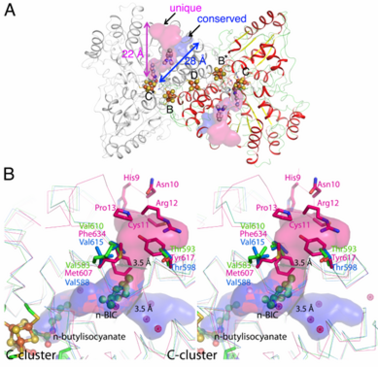
| - | <scene name='Journal:JBIC:13/Cv1/3'>The n-BIC molecule is located in a distance of approximately 8 A from the n-butylisocyanate bound in the active site, measured between nearest atoms and is placed within a hydrophobic channel</scene>, which was shown to be a binding place for Xe atoms in CODH/ACS ([[2z8y]]). Analysis of the CODH-II structure identified the presence of two different channels, in which one is analogous to the channel identified by Xe-soaking in bifunctional CODH/ACS (conserved channel), while the other seems to be specific for monofunctional CODH (unique channel). The conserved channel in CODH-II is similar in position to that in bifunctional CODH, where the channel is extended to reach the A-cluster of ACS. Monofunctional CODHs have smaller side chains like Thr and Val, while Tyr617 and Phe634 block a passage of the unique channel to the protein surface in bifunctional CODH/ACS. Furthermore, the unique channel is completely blocked by residue His9 – Pro13 to prevent diffusion of gaseous substrate/product from the protein in bifunctional CODH/ACS. However, the channel of monofunctional CODH-II is directed towards the solvent, which is in line with its role to allow fast progress and egress of substrates and products from the active site to the outside of the enzyme, which has not been described previously. | + | <scene name='Journal:JBIC:13/Cv1/3'>The n-BIC molecule is located in a distance of approximately 8 A from the n-butylisocyanate bound in the active site, measured between nearest atoms and is placed within a hydrophobic channel</scene>, which was shown to be a binding place for Xe atoms in CODH/ACS ([[2z8y]]). Analysis of the CODH-II structure (see below)identified the presence of two different channels, in which one is analogous to the channel identified by Xe-soaking in bifunctional CODH/ACS (conserved channel), while the other seems to be specific for monofunctional CODH (unique channel). The conserved channel in CODH-II is similar in position to that in bifunctional CODH, where the channel is extended to reach the A-cluster of ACS. Monofunctional CODHs have smaller side chains like Thr and Val, while Tyr617 and Phe634 block a passage of the unique channel to the protein surface in bifunctional CODH/ACS. Furthermore, the unique channel is completely blocked by residue His9 – Pro13 to prevent diffusion of gaseous substrate/product from the protein in bifunctional CODH/ACS. However, the channel of monofunctional CODH-II is directed towards the solvent, which is in line with its role to allow fast progress and egress of substrates and products from the active site to the outside of the enzyme, which has not been described previously. |
| + | [[Image:IM.png|left|378px|thumb|Channels in monofunctional CODH-II<sub>''Ch''</sub>. The channel into which the alkyl group of n-BIC is directed, shown in pink color, is unique to monofunctional CODHs. Blue space-filling model shows the channel found in both mono- and bi-functional CODHs. (B) Comparison of channels in mono- (CODH-II<sub>''Ch''</sub>in green from this study and CODH from ''Rhodospirillum rubrum'' in cyan ([[1jqk]])) and bi-functional (CODH from ''Moorella thermoacetica'' in pink from PDB-ID [[2z8y]]) CODHs. Pink-colored dots are Xenon atoms found in CODH from ''Moorella thermoacetica''.]] | ||
</StructureSection> | </StructureSection> | ||
Revision as of 11:57, 8 September 2011
| |||||||||||
- ↑ 1
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.