Multiple sclerosis
From Proteopedia
| Line 23: | Line 23: | ||
===Interferon Alpha, Interferon Beta, and Interferon Receptors 1 & 2 === | ===Interferon Alpha, Interferon Beta, and Interferon Receptors 1 & 2 === | ||
Since a PDB reference does not exist for interferon beta interacting with interferon receptors 1 or 2, and a multitude of files exist on <scene name='Multiple_sclerosis/Ifna/1'>Interferon-α</scene> interacting with the receptor, a comparison to interferon-α will be made prior to demonstrating the types of bonding that occur between the interferon and its receptor. To see more information regarding interferons, please visit the [[Interferons]] site. | Since a PDB reference does not exist for interferon beta interacting with interferon receptors 1 or 2, and a multitude of files exist on <scene name='Multiple_sclerosis/Ifna/1'>Interferon-α</scene> interacting with the receptor, a comparison to interferon-α will be made prior to demonstrating the types of bonding that occur between the interferon and its receptor. To see more information regarding interferons, please visit the [[Interferons]] site. | ||
| - | |||
Interferon alpha has a 31% sequence homology to interferon beta. It, too, has many <scene name='Multiple_sclerosis/Ifna_labeled/1'>identifiable regions</scene> with two <scene name='Multiple_sclerosis/Ifna_disulfide_bonds/1'>disulfide bonds</scene>: one between the <scene name='Multiple_sclerosis/Ifna_disulfide_bondsn-e/1'>N-terminus and Helix E</scene>, and the other between <scene name='Multiple_sclerosis/Ifna_disulfide_bonds_ab-g/1'>Loop AB and Helix G</scene>. It has seven <scene name='Multiple_sclerosis/Ifna_alphahelices/1'>alpha helices</scene>, as compared to the five of interferon-β, and therefore has several more <scene name='Multiple_sclerosis/Ifna_loops_regions/1'>loop regions.</scene> The helices A, C, and F run <scene name='Multiple_sclerosis/Ifna_parallelacf/2'>parallel</scene> to one another, and <scene name='Multiple_sclerosis/Ifna_antiparallel/1'>anti-parallel</scene> to B, E, and G which run <scene name='Multiple_sclerosis/Ifna_parallel_beg/2'>parallel</scene> to each other. | Interferon alpha has a 31% sequence homology to interferon beta. It, too, has many <scene name='Multiple_sclerosis/Ifna_labeled/1'>identifiable regions</scene> with two <scene name='Multiple_sclerosis/Ifna_disulfide_bonds/1'>disulfide bonds</scene>: one between the <scene name='Multiple_sclerosis/Ifna_disulfide_bondsn-e/1'>N-terminus and Helix E</scene>, and the other between <scene name='Multiple_sclerosis/Ifna_disulfide_bonds_ab-g/1'>Loop AB and Helix G</scene>. It has seven <scene name='Multiple_sclerosis/Ifna_alphahelices/1'>alpha helices</scene>, as compared to the five of interferon-β, and therefore has several more <scene name='Multiple_sclerosis/Ifna_loops_regions/1'>loop regions.</scene> The helices A, C, and F run <scene name='Multiple_sclerosis/Ifna_parallelacf/2'>parallel</scene> to one another, and <scene name='Multiple_sclerosis/Ifna_antiparallel/1'>anti-parallel</scene> to B, E, and G which run <scene name='Multiple_sclerosis/Ifna_parallel_beg/2'>parallel</scene> to each other. | ||
<scene name='Multiple_sclerosis/Ifna_notparalleltoanyoned/1'>Helix D</scene> does not run parallel or anti-parallel to either set, but rather runs at a 45-90 degree angle to them. Helix A consists of residues 10-12; Helix B of 40-43; Helix C of 53-68; Helix D of 70-75; Helix E of 78-100; Helix F of 109-132; and Helix G of 137-158. | <scene name='Multiple_sclerosis/Ifna_notparalleltoanyoned/1'>Helix D</scene> does not run parallel or anti-parallel to either set, but rather runs at a 45-90 degree angle to them. Helix A consists of residues 10-12; Helix B of 40-43; Helix C of 53-68; Helix D of 70-75; Helix E of 78-100; Helix F of 109-132; and Helix G of 137-158. | ||
| - | |||
Interferons -α and -β interact with a receptor at the cell surface.<ref>[http://www.jbc.org/content/282/28/20045.full?sid=cbf08059-44d4-4957-8ea7-0351cab9c2ac] Samuel, C.E. "Interferons, Interferon Receptors, Signal Transducer and Transcriptional Activators, and Inteferon Regulatory Factors." ''J Biol Chem'' 2007 282: 20045-20046. First Published on May 14, 2007, doi:10.1074/jbc.R700025200</ref> This receptor has <scene name='Multiple_sclerosis/Ifnr_domains_labeled/1'>three domains</scene>: an | Interferons -α and -β interact with a receptor at the cell surface.<ref>[http://www.jbc.org/content/282/28/20045.full?sid=cbf08059-44d4-4957-8ea7-0351cab9c2ac] Samuel, C.E. "Interferons, Interferon Receptors, Signal Transducer and Transcriptional Activators, and Inteferon Regulatory Factors." ''J Biol Chem'' 2007 282: 20045-20046. First Published on May 14, 2007, doi:10.1074/jbc.R700025200</ref> This receptor has <scene name='Multiple_sclerosis/Ifnr_domains_labeled/1'>three domains</scene>: an | ||
<scene name='Multiple_sclerosis/Ifnr_n_domain_labeled/1'>N-domain, with two disulfide bonds</scene>, a <scene name='Multiple_sclerosis/Ifnr_c_domain_labeled/1'>C-domain, with one disulfide bond</scene>, and a <scene name='Multiple_sclerosis/Ifnr_linker_region_labeled/1'>linker region</scene>. The <scene name='Multiple_sclerosis/Ifnr_termini_labeled/1'>termini regions</scene> of the receptor have no secondary structure, allowing for some serious flexibility, leading to <scene name='Multiple_sclerosis/Ifnr_clash_n-c/1'>eight clashes amongst the domains</scene>.<ref name="Interferon Receptor Structure">PMID:12842042</ref> | <scene name='Multiple_sclerosis/Ifnr_n_domain_labeled/1'>N-domain, with two disulfide bonds</scene>, a <scene name='Multiple_sclerosis/Ifnr_c_domain_labeled/1'>C-domain, with one disulfide bond</scene>, and a <scene name='Multiple_sclerosis/Ifnr_linker_region_labeled/1'>linker region</scene>. The <scene name='Multiple_sclerosis/Ifnr_termini_labeled/1'>termini regions</scene> of the receptor have no secondary structure, allowing for some serious flexibility, leading to <scene name='Multiple_sclerosis/Ifnr_clash_n-c/1'>eight clashes amongst the domains</scene>.<ref name="Interferon Receptor Structure">PMID:12842042</ref> | ||
| - | Interferon-α <scene name='Multiple_sclerosis/Ifnawithreceptorcolored/1'>binds</scene> to an interferon receptor mainly with helices C and G. There are many <scene name='Multiple_sclerosis/Ifnawithreceptorintrxns/2'>residues</scene> within 4 angstroms of one another. These residues could form many <scene name='Multiple_sclerosis/Ifnawithreceptorintrxns/5'>different types of bonds</scene>, illustrated in white dotted lines. | + | Interferon-α <scene name='Multiple_sclerosis/Ifnawithreceptorcolored/1'>binds</scene> to an interferon receptor mainly with helices C and G. There are many <scene name='Multiple_sclerosis/Ifnawithreceptorintrxns/2'>residues</scene> within 4 angstroms of one another. These residues could form many <scene name='Multiple_sclerosis/Ifnawithreceptorintrxns/5'>different types of bonds</scene>, illustrated in white dotted lines. Given that interferon-α does not undergo many structural changes upon binding to interferon receptor II, Quadt-Akabayov et al. have concluded that the binding mechanism is similar to that of a lock and key. |
===Interferon Beta and MS=== | ===Interferon Beta and MS=== | ||
Revision as of 15:32, 22 April 2012
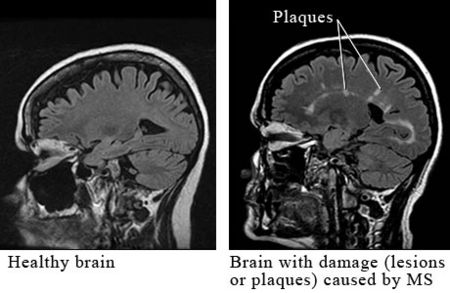
Multiple sclerosis (MS) - an autoimmune disease that effects every patient differently based on the neurologic lesions inflicted throughout the body. While some can go through their lives with relatively mild symptoms and short periods of relapse, others can become incapacitated within years or even months. Defined by Nylander and Hafler, MS is a "multifocal demyelinating disease with progressive neurodegeneration caused by an autoimmune response to self-antigens in a genetically susceptible individual."[2] Inflammation is the primary cause of damage in MS, and though the effects of the disease are well known, and various treatments exist for the disease, the exact identity of an antigen or infectious agent that causes the initiation of a myriad of symptoms is unknown.[3]
There are three ways in which MS is categorized: relapsing-remitting (RRMS), secondary progressive (SPMS), and primary progressive (PPMS). In RRMS, the patient experiences periods of time in which the symptoms increase considerably, although the neurological function of the patient usually returns to normal after the episode. Those with SPMS have symptoms like RRMS, but do not return to normal neurological function after the episode, rather they sustain the neurological damage (such as permanently losing the use of an arm). In PPMS, the patient has an initial episode that never ends. That is, once the symptoms begin, there is no remission in the neurological degradation. A constant autoimmune attack on the patient's body causes increasingly severe symptoms, which can sometimes lead to death.
Taking a biochemical look at the immunopathology and some of the various treatments that exist for MS helps in the understanding that MS is no longer a diagnosis which is hopeless, but is in fact full of hopeful and helpful treatments.
| |||||||||||
References
- ↑ [1] Poinier, A.C., Husney, A., and Chalk, C. "Magnetic resonance imaging (MRI) of multiple sclerosis." Health.com Updated: 2010 Feb 18.
- ↑ 2.0 2.1 2.2 Nylander A, Hafler DA. Multiple sclerosis. J Clin Invest. 2012 Apr 2;122(4):1180-8. doi: 10.1172/JCI58649. Epub 2012 Apr 2. PMID:22466660 doi:10.1172/JCI58649
- ↑ Loma I, Heyman R. Multiple sclerosis: pathogenesis and treatment. Curr Neuropharmacol. 2011 Sep;9(3):409-16. PMID:22379455 doi:10.2174/157015911796557911
- ↑ Dziedzic T, Metz I, Dallenga T, Konig FB, Muller S, Stadelmann C, Bruck W. Wallerian degeneration: a major component of early axonal pathology in multiple sclerosis. Brain Pathol. 2010 Sep;20(5):976-85. Epub 2010 Apr 14. PMID:20477831 doi:10.1111/j.1750-3639.2010.00401.x
- ↑ Smith KJ, Lassmann H. The role of nitric oxide in multiple sclerosis. Lancet Neurol. 2002 Aug;1(4):232-41. PMID:12849456
- ↑ Campbell GR, Ziabreva I, Reeve AK, Krishnan KJ, Reynolds R, Howell O, Lassmann H, Turnbull DM, Mahad DJ. Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann Neurol. 2011 Mar;69(3):481-92. doi: 10.1002/ana.22109. Epub 2010 Nov 8. PMID:21446022 doi:10.1002/ana.22109
- ↑ Mosyak L, Wood A, Dwyer B, Buddha M, Johnson M, Aulabaugh A, Zhong X, Presman E, Benard S, Kelleher K, Wilhelm J, Stahl ML, Kriz R, Gao Y, Cao Z, Ling HP, Pangalos MN, Walsh FS, Somers WS. The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J Biol Chem. 2006 Nov 24;281(47):36378-90. Epub 2006 Sep 27. PMID:17005555 doi:M607314200
- ↑ Voet, D., Voet, J.G., and C. Pratt. Fundamentals of Biochemistry 3rd Edition. Hoboken, NJ: John Wiley and Sons, 2008. Print.
- ↑ Kudo M. Management of hepatocellular carcinoma: from prevention to molecular targeted therapy. Oncology. 2010 Jul;78 Suppl 1:1-6. Epub 2010 Jul 8. PMID:20616576 doi:10.1159/000315222
- ↑ http://www.uniprot.org/uniprot/P00784
- ↑ [2] Samuel, C.E. "Interferons, Interferon Receptors, Signal Transducer and Transcriptional Activators, and Inteferon Regulatory Factors." J Biol Chem 2007 282: 20045-20046. First Published on May 14, 2007, doi:10.1074/jbc.R700025200
- ↑ Chill JH, Quadt SR, Levy R, Schreiber G, Anglister J. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure. 2003 Jul;11(7):791-802. PMID:12842042
Relevant 3D Structures
Interferon Beta
1au1 - Homo sapiens
Interferon Receptors
3s98, 3se3, 3se4, 1n6u, 1n6v, 2hym, 2kz1, 2lag, 3s8w, 3s9d - Homo sapiens
