Interferon
From Proteopedia
| Line 16: | Line 16: | ||
===Interferon-β=== | ===Interferon-β=== | ||
A protein growth factor that stimulates an antiviral defense <scene name='Multiple_sclerosis/Interferon_beta/9'>interferon-beta</scene> is one of the only two known vertebrate structural genes that lacks introns.<ref name="Biochem Text">Voet, D., Voet, J.G., and C. Pratt. ''Fundamentals of Biochemistry'' 3rd Edition. Hoboken, NJ: John Wiley and Sons, 2008. Print.</ref> Interferon-β has a 31% sequence homology to interferon-α . It is a relatively simple biological response modifier, with several <scene name='Multiple_sclerosis/Interferon_beta_labeled/1'>identifiable regions</scene>. It consists of five <scene name='Multiple_sclerosis/Ifnb_helices_in_color/1'>alpha helices</scene>, as compared to the seven of interferon-α, as well as multiple interconnecting <scene name='Multiple_sclerosis/Interferon_beta_loops/2'>loop regions</scene>. Helices A, B and D run <scene name='Multiple_sclerosis/Ifnb_parallel_abd/3'>parallel to one another</scene>, and helices C and E run <scene name='Multiple_sclerosis/Ifnb_antiparallel/1'>anti-parallel</scene> to the other three helices, but <scene name='Multiple_sclerosis/Ifnb_antiparallel_ce/3'>parallel</scene> to one another. Helix A consists of residues 6-23; Helix B consists of residues 49-65; Helix C consists of residues 77-91; Helix D consists of residues 112-131; and Helix E consists of residues 135-155.<ref name="Structure Ifn B">PMID:20616576</ref><ref name="UniProt">http://www.uniprot.org/uniprot/P00784</ref> | A protein growth factor that stimulates an antiviral defense <scene name='Multiple_sclerosis/Interferon_beta/9'>interferon-beta</scene> is one of the only two known vertebrate structural genes that lacks introns.<ref name="Biochem Text">Voet, D., Voet, J.G., and C. Pratt. ''Fundamentals of Biochemistry'' 3rd Edition. Hoboken, NJ: John Wiley and Sons, 2008. Print.</ref> Interferon-β has a 31% sequence homology to interferon-α . It is a relatively simple biological response modifier, with several <scene name='Multiple_sclerosis/Interferon_beta_labeled/1'>identifiable regions</scene>. It consists of five <scene name='Multiple_sclerosis/Ifnb_helices_in_color/1'>alpha helices</scene>, as compared to the seven of interferon-α, as well as multiple interconnecting <scene name='Multiple_sclerosis/Interferon_beta_loops/2'>loop regions</scene>. Helices A, B and D run <scene name='Multiple_sclerosis/Ifnb_parallel_abd/3'>parallel to one another</scene>, and helices C and E run <scene name='Multiple_sclerosis/Ifnb_antiparallel/1'>anti-parallel</scene> to the other three helices, but <scene name='Multiple_sclerosis/Ifnb_antiparallel_ce/3'>parallel</scene> to one another. Helix A consists of residues 6-23; Helix B consists of residues 49-65; Helix C consists of residues 77-91; Helix D consists of residues 112-131; and Helix E consists of residues 135-155.<ref name="Structure Ifn B">PMID:20616576</ref><ref name="UniProt">http://www.uniprot.org/uniprot/P00784</ref> | ||
| + | |||
| + | Interferon-β is used as a treatment for [[Multiple sclerosis]], an autoimmune disease defined by Nylander and Hafler as "a multifocal demyelinating disease with progressive neurodegeneration caused by an autoimmune response to self-antigens in a genetically susceptible individual."<ref name ="MS Nylander & Hafler">PMID:22466660</ref> Inflammation is the primary cause of damage in MS, and though the effects of the disease are well known and various treatments exist for the disease, the exact identity of an antigen or infectious agent that causes the initiation of a myriad of symptoms is unknown.<ref name='MS:Pathogenesis and Treatment'>PMID:22379455</ref> | ||
__NOTOC__ | __NOTOC__ | ||
Revision as of 00:15, 25 April 2012
Interferons were the first cytokines discovered and were identified by Isaacs and Lindenmann. These proteins were classified as interferons because they interfered with virus growth.[1] The initial experiments performed poorly characterized the interferons, and was based merely on bioactivity. Advances in scientific instrumentation and technique have allowed for greater understanding and visualization of not only the structure but also the mechanisms of the various types of inteferons.[2] The interferons were originally classified as leukocyte (interferon-α), fibroblast (interferon-β), and immmune (interferon-γ), although today they are classified into types I (α, β, ε, κ, ω), II (γ), and III (λ).[1][2]
| |||||||||||
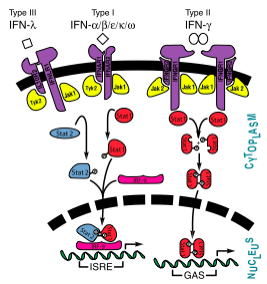
Signaling and Receptor Interactions
The signaling pathways of interferons are interesting as type I interferons share the same receptors IFNAR1 and IFNAR2. Type II interferon-γ has receptors IFNGR1 and IFNGR2, but needs two interferon-γ to signal, as illustrated in the image to the right. Interestingly enough, types I and III act together in the JAK-STAT pathway, while type II acts alone. Interferon-α and -β bind to the same receptors as one another, the affinities with which they bind to IFNAR1 and IFNAR2 differ. While the binding to IFNAR2 is stronger for both in comparison to IFNAR1, interferon-β has a much stronger affinity for IFNAR1 than interferon-α.[9]
Interferon-α to an interferon receptor mainly with helices C and G. There are many within 4 angstroms of one another. These residues could form many , illustrated in white dotted lines. Given that interferon-α does not undergo many structural changes upon binding to interferon receptor II, Quadt-Akabayov et al. have concluded that the binding mechanism is similar to that of a lock and key. Interferons -α and -β interact with a receptor at the cell surface.[10] This receptor has : an , with two disulfide bonds, a , with one disulfide bond, and a . The of the receptor have no secondary structure, allowing for some serious flexibility, leading to .[11]
References
- ↑ 1.0 1.1 1.2 [1] Samuel, C.E. "Interferons, Interferon Receptors, Signal Transducer and Transcriptional Activators, and Inteferon Regulatory Factors." J Biol Chem 2007 282: 20045-20046. First Published on May 14, 2007, doi:10.1074/jbc.R700025200
- ↑ 2.0 2.1 Langer JA, Pestka S. Structure of interferons. Pharmacol Ther. 1985;27(3):371-401. PMID:2413490
- ↑ Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J. Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex. Protein Sci. 2006 Nov;15(11):2656-68. Epub 2006 Sep 25. PMID:17001036 doi:10.1110/ps.062283006
- ↑ Voet, D., Voet, J.G., and C. Pratt. Fundamentals of Biochemistry 3rd Edition. Hoboken, NJ: John Wiley and Sons, 2008. Print.
- ↑ Kudo M. Management of hepatocellular carcinoma: from prevention to molecular targeted therapy. Oncology. 2010 Jul;78 Suppl 1:1-6. Epub 2010 Jul 8. PMID:20616576 doi:10.1159/000315222
- ↑ http://www.uniprot.org/uniprot/P00784
- ↑ Nylander A, Hafler DA. Multiple sclerosis. J Clin Invest. 2012 Apr 2;122(4):1180-8. doi: 10.1172/JCI58649. Epub 2012 Apr 2. PMID:22466660 doi:10.1172/JCI58649
- ↑ Loma I, Heyman R. Multiple sclerosis: pathogenesis and treatment. Curr Neuropharmacol. 2011 Sep;9(3):409-16. PMID:22379455 doi:10.2174/157015911796557911
- ↑ Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J. Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex. Protein Sci. 2006 Nov;15(11):2656-68. Epub 2006 Sep 25. PMID:17001036 doi:10.1110/ps.062283006
- ↑ [2] Samuel, C.E. "Interferons, Interferon Receptors, Signal Transducer and Transcriptional Activators, and Inteferon Regulatory Factors." J Biol Chem 2007 282: 20045-20046. First Published on May 14, 2007, doi:10.1074/jbc.R700025200
- ↑ Chill JH, Quadt SR, Levy R, Schreiber G, Anglister J. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure. 2003 Jul;11(7):791-802. PMID:12842042
Relevant 3D Structures
Interferon-α
Interferon-β
1au1 - Homo sapiens
Inteferon-α/β Receptors
Interferon-α/β Receptors in Complex with Interferon-α
3oq3, 3se3, 3se4, 1n6u, 1n6v, 2hym, 2kz1, 2lag, 3s9d - Homo sapiens
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Kirsten Eldredge, Alexander Berchansky, Joel L. Sussman, Karl Oberholser, Jaime Prilusky
