Sandbox Reserved 482
From Proteopedia
| Line 3: | Line 3: | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
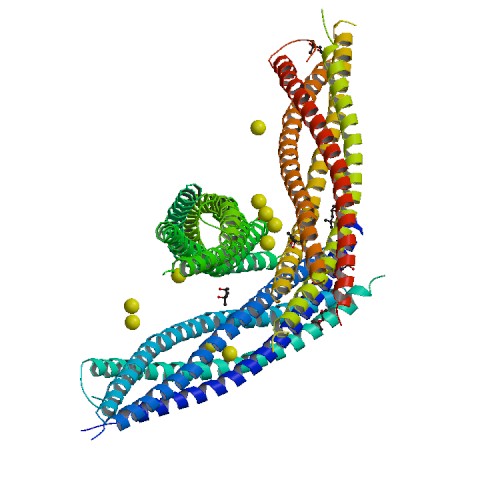
| - | <Structure load='1KIL' size='500' frame='true' align='right' caption='Syntaxin-1A - Three-dimensional structure of the complexin/SNARE complex.' scene='Insert optional scene name here' /> | + | <Structure load='1KIL' size='500' frame='true' align='right' caption='Syntaxin-1A - Three-dimensional structure of the complexin/SNARE complex, X-RAY DIFFRACTION with resolution of 2.30 Å.' scene='Insert optional scene name here' /> |
== Syntaxin-1A == | == Syntaxin-1A == | ||
| Line 26: | Line 26: | ||
| - | NMR spectroscopy was used to identify a folded N-terminal domain in Syntaxin-1A and to determine its three-dimensional structure. This 120 residue N-terminal domain is conserved in plasma membrane syntaxins. The domain is made up from three long <scene name='Sandbox_Reserved_482/Syntaxin-1a/6'>alpha helices</scene> and an up-and-down bundle with a left-handed twist. (4) | + | NMR spectroscopy was used to identify a folded N-terminal domain in Syntaxin-1A and to determine its three-dimensional structure. This 120 residue N-terminal domain is conserved in plasma membrane syntaxins. The domain is made up from three long <scene name='Sandbox_Reserved_482/Syntaxin-1a/6'>alpha helices</scene> and an up-and-down bundle with a left-handed twist. Two <scene name='Sandbox_Reserved_482/Syntaxin-1a/7'>ligands</scene> are displayed within the SNARE complex. (4) |
The image below shows a neuronal synaptic fusion complex to exemplify the use of the <scene name='Sandbox_Reserved_482/Syntaxin-1a/2'>active site</scene>, ASP 68. SNARE proteins are involved in the fusion of vesicles with their target membranes but the structural details of these complexes are still unknown. X-ray is here used to show the crystal structure at 2.4 Å resolution of a core synaptic fusion complex containing syntaxin-1A, synaptobrevin-II and SNAP-25B. (6) | The image below shows a neuronal synaptic fusion complex to exemplify the use of the <scene name='Sandbox_Reserved_482/Syntaxin-1a/2'>active site</scene>, ASP 68. SNARE proteins are involved in the fusion of vesicles with their target membranes but the structural details of these complexes are still unknown. X-ray is here used to show the crystal structure at 2.4 Å resolution of a core synaptic fusion complex containing syntaxin-1A, synaptobrevin-II and SNAP-25B. (6) | ||
Revision as of 22:16, 28 April 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | ||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
Syntaxin-1AIntroduction Syntaxin-1A is part of the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) family. It is an integrin plasma membrane protein found almost exclusively in neurons and it is known for its essential roles in neuronal signaling. (1) It assembles in tight core complexes, which promote fusion of carrier vesicles with target compartments. Members of the SNARE class of proteins are expressed in all eukaryotic cells and are distributed in distinct subcellular compartments. (2)
Syntaxin-1A has 288 residues. Residue 1-265 make up the cytoplasmic domain and residue 266-288 form the carboxyl-terminal transmembrane anchor. It adapts a well-folded tertiary structure with repeated forming a colied coil structure. (5)
The image below shows a neuronal synaptic fusion complex to exemplify the use of the , ASP 68. SNARE proteins are involved in the fusion of vesicles with their target membranes but the structural details of these complexes are still unknown. X-ray is here used to show the crystal structure at 2.4 Å resolution of a core synaptic fusion complex containing syntaxin-1A, synaptobrevin-II and SNAP-25B. (6)
Syntaxin-1A has an essential role in neuronal signaling. It regulates fusion, docking and trafficking of synaptic vesicles with the plasma membrane. The protein also plays an important role in internalization of vesicles containing a transporter for the neurotransmitter glutamate. The transportation of glutamate is important to avoid excessive stimulation of glutamate receptors and to support the conversion to the inhibitory neurotransmitter GABA. Syntaxin-1A also regulates ion channels such as Ca2+ channels, K+ channels and epithelial Na+ channels. (7) Applications |