We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 488
From Proteopedia
(Difference between revisions)
(→Background) |
|||
| Line 15: | Line 15: | ||
== Structure == | == Structure == | ||
| - | <Structure load='1A93' size='500' frame='true' align='right' caption='NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE' scene='Insert optional scene name here' /> | + | <Structure load='1A93' size='500' frame='true' align='right' caption='NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE <ref name="PDB">PMID:9680483</ref>' scene='Insert optional scene name here' /> |
The c-myc gene is consisted of three exons located on human chromosomes 8q24. The c-Myc sequence contains many Myc boxes, or conserved N-terminal domains, which are found in N-Myc and L-Myc.<ref name="cancer mechanism" /> The C-terminal region of c-Myc contains the helix-loop-helix, leucine zipper, and a basic region. These three components dimerize to form a bHLH LZ motif. The binding of DNA is achieved by the combination of the bHLH LZ motif and another bHLH LZ protein, Max.DNA-bound Myc-Max complexes activate transcription through the amino terminal 143 amino acids of c-Myc, known as the transcriptional activation domain (TAD). <ref name="Cancer" /> | The c-myc gene is consisted of three exons located on human chromosomes 8q24. The c-Myc sequence contains many Myc boxes, or conserved N-terminal domains, which are found in N-Myc and L-Myc.<ref name="cancer mechanism" /> The C-terminal region of c-Myc contains the helix-loop-helix, leucine zipper, and a basic region. These three components dimerize to form a bHLH LZ motif. The binding of DNA is achieved by the combination of the bHLH LZ motif and another bHLH LZ protein, Max.DNA-bound Myc-Max complexes activate transcription through the amino terminal 143 amino acids of c-Myc, known as the transcriptional activation domain (TAD). <ref name="Cancer" /> | ||
Revision as of 23:21, 2 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | ||||||||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
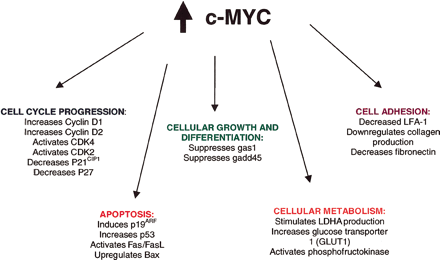
c-Myc ProteinBackgroundc-Myc protein, also known as Transcription factor p64, is a product of the c-myc proto-oncogene that has the ability to bind to DNA and regulate gene transcription.[1] More specifically, c-Myc recognizes the DNA's core sequence 5'-CAC[GA]TG-3' by forming a heterodimer with MAX, a bHLh protein. This structure then interact with TAF1C, SPAG9,PARP10, KDM5A, and KDM5B. This protein is found mainly in Human and it's known to express the MYC gene using an E.coli expression host. [2] Structure
|
||||||||||||