Sandbox Reserved 482
From Proteopedia
| Line 45: | Line 45: | ||
| - | Noradrenaline transporters (NETs) terminate noradrenergic transmission and are often used as a target for antidepressant medications. Syntaxin-1A interacts with NET and modulates NET activity. Protein kinase C activation disrupts NET/syntaxin-1A interactions and down regulates NET activity. Syntaxin-1A binds the NH2 terminal of NET. The protein both supports the surface trafficking of NET and limits the transporter function through its direct interaction with NET. The net result of these two factors is a dynamic cycle of interactions that leads to coordinated control between noradrenaline release and uptake. ( | + | Noradrenaline transporters (NETs) terminate noradrenergic transmission and are often used as a target for antidepressant medications. Syntaxin-1A interacts with NET and modulates NET activity. Protein kinase C activation disrupts NET/syntaxin-1A interactions and down regulates NET activity. Syntaxin-1A binds the NH2 terminal of NET. The protein both supports the surface trafficking of NET and limits the transporter function through its direct interaction with NET. The net result of these two factors is a dynamic cycle of interactions that leads to coordinated control between noradrenaline release and uptake.<ref>Sung, Uhna, Aurelio Galli, Kristopher Kahlig, et al. "A Regulated Interaction of Syntaxin 1A with the Antidepressant-Sensitive Norepinephrine Transporter Establishes Catecholamine Clearance Capacity." Journal of Neuroscience. 23.5 (2003): 1697-1709. Web. 28 May. 2012. <http://www.jneurosci.org/content/23/5/1697.full></ref> |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 02:14, 3 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
Syntaxin-1AIntroductionSyntaxin-1A is part of the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) family. It is an integrin plasma membrane protein found almost exclusively in neurons and it is known for its essential roles in neuronal signaling. [1] It assembles in tight core complexes, which promote fusion of carrier vesicles with target compartments. Members of the SNARE class of proteins are expressed in all eukaryotic cells and are distributed in distinct subcellular compartments.
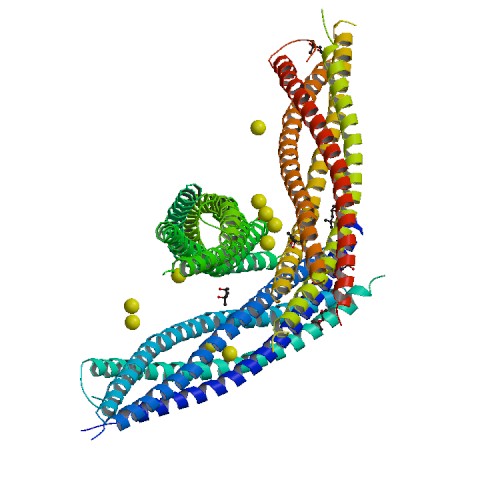
StructureSyntaxin-1A has 288 residues. Residue 1-265 make up the cytoplasmic domain and residue 266-288 form the carboxyl-terminal transmembrane anchor. It adapts a well-folded tertiary structure with repeated forming a colied coil structure.[3]
The image below shows a neuronal synaptic fusion complex to exemplify the use of the , ASP 68. SNARE proteins are involved in the fusion of vesicles with their target membranes but the structural details of these complexes are still unknown. X-ray is here used to show the crystal structure at 2.4 Å resolution of a core synaptic fusion complex containing syntaxin-1A, synaptobrevin-II and SNAP-25B.[5]
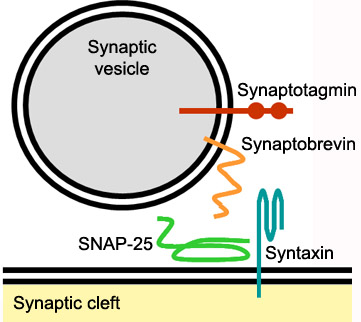
Mechanism of actionSyntaxin-1A has an essential role in neuronal signaling. It regulates fusion, docking and trafficking of synaptic vesicles with the plasma membrane. The SNARE complex is a four-alpha-helix bundle composed of syntaxin-1A, synaptobrevin and SNAP-25. In neuronal exocytosis, syntaxin and synaptobrevin are attached in respective membranes by their C-terminal domains, whereas SNAP-25 is connected to the plasma membrane via several cysteine-linked palmitoyl chains. Synaptotagmin acts as a calcium sensor and regulates the SNARE closure. The influx of calcium into the cell triggers the completion of the assembly reaction.
ApplicationsAs mentioned before, the neurodevelopmental disorder Williams syndrome is related to the deletion of, among others, syntaxin-1A. Other typical genes that are deleted are CLIP2, ELN, GTF2I, GTF2IRD1, and LIMK1. The most common symptoms of Williams syndrome are mental disability, heart defects, and unusual facial features.[7]
References
|